Regulation of gene expression in eukaryotes
Regulation of gene expression in eukaryotes
Regulation of gene expression in eukaryotes
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
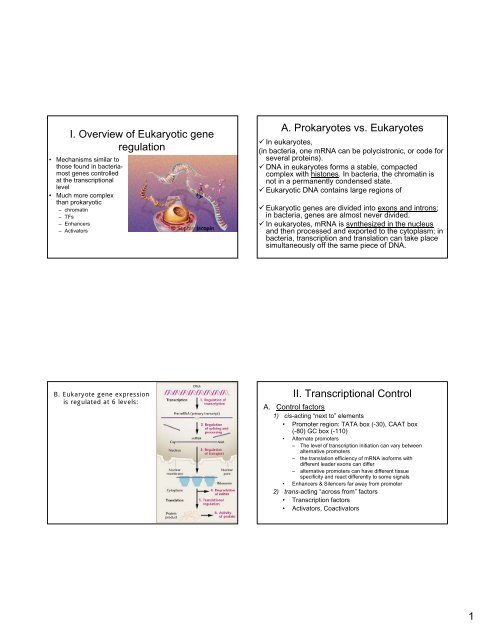
I. Overview <strong>of</strong> Eukaryotic <strong>gene</strong><br />
regulation<br />
• Mechanisms similar to<br />
those found <strong>in</strong> bacteriamost<br />
<strong>gene</strong>s controlled<br />
at the transcriptional<br />
level<br />
• Much more complex<br />
than prokaryotic<br />
– chromat<strong>in</strong><br />
– TFs<br />
– Enhancers<br />
– Activators<br />
A. Prokaryotes vs. Eukaryotes<br />
In <strong>eukaryotes</strong>,<br />
(<strong>in</strong> bacteria, one mRNA can be polycistronic, or code for<br />
several prote<strong>in</strong>s).<br />
DNA <strong>in</strong> <strong>eukaryotes</strong> forms a stable, compacted<br />
complex with histones. In bacteria, the chromat<strong>in</strong> is<br />
not <strong>in</strong> a permanently condensed state.<br />
Eukaryotic DNA conta<strong>in</strong>s large regions <strong>of</strong><br />
Eukaryotic <strong>gene</strong>s are divided <strong>in</strong>to exons and <strong>in</strong>trons;<br />
<strong>in</strong> bacteria, <strong>gene</strong>s are almost never divided.<br />
In <strong>eukaryotes</strong>, mRNA is synthesized <strong>in</strong> the nucleus<br />
and then processed and exported to the cytoplasm; <strong>in</strong><br />
bacteria, transcription and translation can take place<br />
simultaneously <strong>of</strong>f the same piece <strong>of</strong> DNA.<br />
B. Eukaryote <strong>gene</strong> <strong>expression</strong><br />
is regulated at 6 levels:<br />
II. Transcriptional Control<br />
A. Control factors<br />
1) cis-act<strong>in</strong>g “next to” elements<br />
• Promoter region: TATA box (-30), CAAT box<br />
(-80) GC box (-110)<br />
• Alternate promoters<br />
– The level <strong>of</strong> transcription <strong>in</strong>itiation can vary between<br />
alternative promoters<br />
– the translation efficiency <strong>of</strong> mRNA is<strong>of</strong>orms with<br />
different leader exons can differ<br />
– alternative promoters can have different tissue<br />
specificity and react differently to some signals<br />
• Enhancers & Silencers far away from promoter<br />
2) trans-act<strong>in</strong>g “across from” factors<br />
• Transcription factors<br />
• Activators, Coactivators<br />
1
Control factors cont<strong>in</strong>ued:<br />
3) DNA methylation (add methyl to C)<br />
• Occurs at 5’ position, usually <strong>in</strong> CG doublets<br />
• 5’- m CpG-3’<br />
– Transcriptionally active <strong>gene</strong>s possess<br />
significantly lower levels <strong>of</strong> methylated DNA<br />
than <strong>in</strong>active <strong>gene</strong>s.<br />
• A <strong>gene</strong> for methylation is essential for development <strong>in</strong><br />
mice (turn<strong>in</strong>g <strong>of</strong>f a <strong>gene</strong> also can be important).<br />
• Methylation results <strong>in</strong> a human disease called fragile<br />
X syndrome; FMR-1 <strong>gene</strong> is silenced by methylation.<br />
Control factors cont<strong>in</strong>ued:<br />
4) Chromat<strong>in</strong> conformation (remodell<strong>in</strong>g)<br />
a. Antirepressors & nucleosome position<strong>in</strong>g.<br />
b. Histone acetylation – (acetyl groups on lys<strong>in</strong>es),<br />
histone acetyltransferase enzyme catalyzes the<br />
addition <strong>of</strong> lys<strong>in</strong>e, targeted to <strong>gene</strong>s by specific<br />
TFs.<br />
c. Heterochromat<strong>in</strong> – highly condensed,<br />
transcriptionally <strong>in</strong>ert (<strong>of</strong>f).<br />
2
B. Eukaryotic Promoters<br />
Usually located with<strong>in</strong> 100 bp upstream<br />
‣ Recognized byRNA Pol II (transcribes mRNA)<br />
‣ Require the b<strong>in</strong>d<strong>in</strong>g <strong>of</strong> several prote<strong>in</strong> factors to <strong>in</strong>itiate<br />
transcription (DNA b<strong>in</strong>d<strong>in</strong>g doma<strong>in</strong>s on TFs – ‘motifs’)<br />
‣ May be positively or negatively regulated<br />
C. Transcription Factors –the<br />
transcription complex<br />
1) TFIIA, TFIIB,<br />
TFIID, TFIIE,<br />
TFIIH<br />
2) TATA b<strong>in</strong>d<strong>in</strong>g<br />
prote<strong>in</strong> (TBP)<br />
3) TBP associated<br />
factors (TAFs)<br />
‣Assembly <strong>of</strong> the basal<br />
transcription apparatus -<br />
<strong>in</strong>volves stepwise b<strong>in</strong>d<strong>in</strong>g<br />
<strong>of</strong> various transcription<br />
factor prote<strong>in</strong>s.<br />
Commitment Stage &<br />
Clearance Stage…<br />
3
Enhancers<br />
Cis regulators that <strong>in</strong>teract<br />
with regulatory prote<strong>in</strong>s &<br />
TFs to <strong>in</strong>crease the<br />
efficiency <strong>of</strong> transcription<br />
<strong>in</strong>itiation.<br />
Silencers – cis-act<strong>in</strong>g,<br />
bound by repressors, or<br />
cause the chromat<strong>in</strong> to<br />
condense and become<br />
<strong>in</strong>active.<br />
D. An example <strong>of</strong> transcriptional control:<br />
Galactose metabolism <strong>in</strong> yeast<br />
‣ GAL1, GAL7, GAL10 <strong>gene</strong>s… products<br />
required for conversion <strong>of</strong> galactose <strong>in</strong>to<br />
glucose<br />
Activators - Prote<strong>in</strong>s that<br />
function by contact<strong>in</strong>g basal<br />
transcription factors and<br />
stimulat<strong>in</strong>g the assembly <strong>of</strong><br />
pre-<strong>in</strong>itiation complexes at<br />
promoters.<br />
Galactose metaboliz<strong>in</strong>g<br />
pathway <strong>of</strong> yeast.<br />
Controll<strong>in</strong>g GAL<br />
GAL80 encodes a prote<strong>in</strong> that negatively regulates<br />
transcription. The repressor prote<strong>in</strong> b<strong>in</strong>ds to an<br />
Activator prote<strong>in</strong>, render<strong>in</strong>g it <strong>in</strong>active.<br />
GAL4 encodes an activator w/z<strong>in</strong>c f<strong>in</strong>ger motif that<br />
activates transcription <strong>of</strong> the three GAL <strong>gene</strong>s<br />
<strong>in</strong>dividually.<br />
Galactose = Inducer, that b<strong>in</strong>ds to Gal80, caus<strong>in</strong>g<br />
it to release Gal4<br />
‣ Although this looks similar to Lac Operon, there<br />
are different molecular mechanisms…<br />
4
Activation model <strong>of</strong> GAL <strong>gene</strong>s <strong>in</strong> yeast.<br />
III. Post-transcriptional control<br />
Alternative polyadenylation and splic<strong>in</strong>g <strong>of</strong> the human CACL <strong>gene</strong> <strong>in</strong><br />
thyroid and neuronal cells.<br />
A. Alternative splic<strong>in</strong>g - Some messages undergo<br />
alternate splic<strong>in</strong>g depend<strong>in</strong>g on what tissue they<br />
are located <strong>in</strong>. The regulation is at the level <strong>of</strong><br />
snRNP production.<br />
– Some pre-mRNAs can be<br />
– Controlled by RNA b<strong>in</strong>d<strong>in</strong>g splic<strong>in</strong>g factors that commit<br />
splic<strong>in</strong>g <strong>in</strong> a particular way<br />
Calciton<strong>in</strong><br />
<strong>gene</strong>-related<br />
peptide<br />
5
Post-transcriptional control cont.<br />
B. The stability <strong>of</strong> a class <strong>of</strong> mRNA can be controlled.<br />
‣ Some short-lived mRNAs have multiple copies <strong>of</strong> the<br />
sequence AUUUA which may act as a target for<br />
degradation.<br />
‣ the hormone prolact<strong>in</strong> enhances the stability <strong>of</strong> the<br />
mRNA for the milk prote<strong>in</strong> case<strong>in</strong><br />
‣ high levels <strong>of</strong> iron decrease the stability <strong>of</strong> the mRNA<br />
for the receptor that br<strong>in</strong>gs iron <strong>in</strong>to cells<br />
C. RNA <strong>in</strong>terference – poorly understood, but<br />
appears to be widespread <strong>in</strong> fungi, plants and<br />
animals as a regulatory mechanism<br />
‣ miRNAs & siRNAS (small RNA molecules) pair with<br />
prote<strong>in</strong>s to form an RNA-<strong>in</strong>duced silenc<strong>in</strong>g complex<br />
(RISC)<br />
‣ RISC pairs w/complentary base sequences <strong>of</strong> specific<br />
mRNAs and causes:<br />
6