Regulation of gene expression in eukaryotes
Regulation of gene expression in eukaryotes
Regulation of gene expression in eukaryotes
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
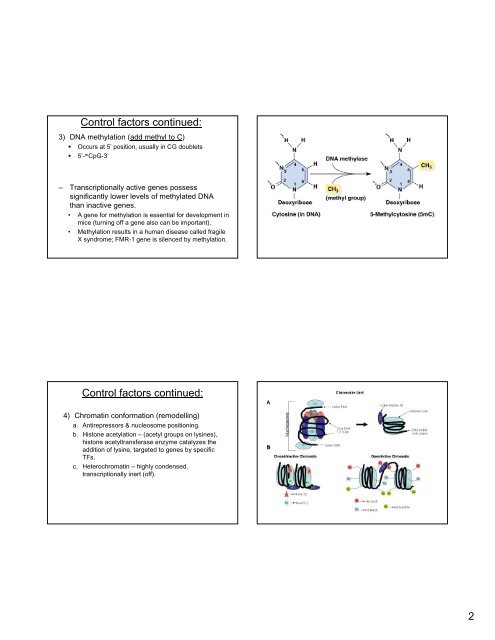
Control factors cont<strong>in</strong>ued:<br />
3) DNA methylation (add methyl to C)<br />
• Occurs at 5’ position, usually <strong>in</strong> CG doublets<br />
• 5’- m CpG-3’<br />
– Transcriptionally active <strong>gene</strong>s possess<br />
significantly lower levels <strong>of</strong> methylated DNA<br />
than <strong>in</strong>active <strong>gene</strong>s.<br />
• A <strong>gene</strong> for methylation is essential for development <strong>in</strong><br />
mice (turn<strong>in</strong>g <strong>of</strong>f a <strong>gene</strong> also can be important).<br />
• Methylation results <strong>in</strong> a human disease called fragile<br />
X syndrome; FMR-1 <strong>gene</strong> is silenced by methylation.<br />
Control factors cont<strong>in</strong>ued:<br />
4) Chromat<strong>in</strong> conformation (remodell<strong>in</strong>g)<br />
a. Antirepressors & nucleosome position<strong>in</strong>g.<br />
b. Histone acetylation – (acetyl groups on lys<strong>in</strong>es),<br />
histone acetyltransferase enzyme catalyzes the<br />
addition <strong>of</strong> lys<strong>in</strong>e, targeted to <strong>gene</strong>s by specific<br />
TFs.<br />
c. Heterochromat<strong>in</strong> – highly condensed,<br />
transcriptionally <strong>in</strong>ert (<strong>of</strong>f).<br />
2