TDS pBLAST42-mcs - InvivoGen
TDS pBLAST42-mcs - InvivoGen
TDS pBLAST42-mcs - InvivoGen
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
<strong>pBLAST42</strong>-<strong>mcs</strong><br />
A plasmid containing the IL-2 signal sequence, a multiple cloning site and the blasticidin resistance gene<br />
Catalog # pbla42-<strong>mcs</strong><br />
For research use only<br />
Version # 05H11-MT<br />
P R O D U C T I N F O R M AT I O N<br />
C o n t e n t :<br />
- 20 µg of lyophilized <strong>pBLAST42</strong>-<strong>mcs</strong> plasmid DNA.<br />
- 4 pouches of E. coli F a s t - M e d i a ® Blast (2 for agar media, 2 for liquid media).<br />
Storage and stability:<br />
- Products are shipped at room temperature.<br />
- Upon receipt, resuspend lyophilized DNA and store at -20°C. Avoid repeated<br />
freeze-thaw cycles.<br />
- Store E. coli F a s t - M e d i a ® Blas at room temperature. Fast-Media ® pouches are<br />
stable 18 months when stored properly.<br />
Quality contro l :<br />
- Plasmid DNA was prepared using an affinity column and lyophilized.<br />
- Plasmid construct has been confirmed by restriction analysis sequencing.<br />
G E N E R A L P R O D U C T U S E<br />
<strong>pBLAST42</strong>-<strong>mcs</strong> is a ready-made expression vector containing the Blasticidin<br />
resistance gene, the hybrid EF1α/ H T LV promoter and a multiple cloning site.<br />
p B L A S T 4 2 - m c s may be used for:<br />
Cloning in a gene of intere s t. Four unique restriction sites comprise the MCS<br />
facilitating cloning of genes. Cloned genes will be under the control of the<br />
E F 1α/ H T LV p r o m o t e r. Genes lacking a native signal sequence will be secreted<br />
as the MCS is located downstream of the human IL-2 signal sequence.<br />
As an “empty” control vector. Since <strong>pBLAST42</strong>-<strong>mcs</strong> does not contain a<br />
therapeutic gene, it can be used in conjunction with other vectors of the pBLAST<br />
family to serve as an experimental control.<br />
p B L A S T is selectable with blasticidin in both E. coli and mammalian cells.<br />
PLASMID FEAT U R E S<br />
• EF-1α / HTLV hybrid pro m o t e r i s a composite promoter comprised of the<br />
Elongation Factor- 1α( E F - 1α) promoter 1 and 5’untranslated region of the Human<br />
T-Cell Leukemia Virus (HTLV). EF-1α is a ‘housekeeping’ gene ubiquitously<br />
expressed in eukaryotic cells. The EF-1α promoter exhibits a strong activity,<br />
higher than viral promoters and, on the contrary to the CMV p r o m o t e r, yields<br />
persistent expression of the transgene in vivo. The R segment and part of the U5<br />
sequence (R-U5’) of the HTLV Type 1 Long Terminal Repeat 2 has been coupled<br />
to the EF-1α promoter to enhance stability of DNA and RNA. This modification<br />
not only increases steady state transcription, but also significantly increases<br />
translation efficiency possibly through mRNA s t a b i l i z a t i o n .<br />
• I117 is a bacterial promoter that is spliced out as an intron in mammalian cells.<br />
Expression of the transgene in E. coli can be further increased by the addition of<br />
IPTG when working with bacteria that constitutively express L a c I<br />
• SV40 polyA: The Simian Virus 40 late polyadenylation signal enables eff i c i e n t<br />
cleavage and polyadenylation reactions resulting in high levels of steady-state<br />
mRNA. The efficiency of this signal was first described by Carswell et al. 3<br />
• SpAn: A synthetic polyadenylation site and a strong pause site are placed<br />
downstream of the pMB1 Ori to limit transcriptional interference between both<br />
transcriptional units. The synthetic polyA site is based on the highly eff i c i e n t<br />
p o l y A signal of the rabbit β-globin gene 4 .<br />
• IL2 ss: The IL2 signal sequence contains 21 amino acids and share common<br />
characteristics with signal peptides of other secretory proteins with respect to<br />
abundance and positions of hydrophobic amino acids. The intracellular cleavage<br />
of the IL2 signal peptide occurs after Ser20 and leads to the secretion of the<br />
expressed protein.<br />
Note: The gene of interest must be cloned in frame with the IL-2ss as translation<br />
will begin at the ATG of the IL-2ss.<br />
• Multiple cloning site.<br />
The MCS contains the restriction sites N r u I, A s c I, B s sH II a n d N h e I.<br />
N ru I is compatible with any blunt-end restriction enzyme.<br />
A s c I is compatible with A f l III, M l u I and B s sH II<br />
B s sH II is compatible with A s c I, A f l III and M l u I .<br />
N h e I is compatible with Xba I, Spe I, and Av r I I .<br />
• pMB1 Ori is a minimal E. coli origin of replication with the same activity as<br />
the longer Ori.<br />
• SV40 prom: The Simian Virus 40 promoter allows the expression of the<br />
blasticidin resistance gene in mammalian cells.<br />
• Bsr (blasticidin resistance gene): The b s r gene from Bacillus cereus e n c o d e s<br />
a deaminase that confers resistance to the antibiotic Blasticidin S. The b s r gene is<br />
driven by the SV40 promoter in tandem with the bacterial EM7 promoter.<br />
Therefore each pBLAST plasmid can be used to select stable mammalian cells<br />
transfectants and E. coli t r a n s f o r m a n t s .<br />
• bGh pAn: The bovine growth hormone (bGh) polyadenylation (pAn) signal<br />
and a transcriptional pause are placed 3' of the blasticidin gene. The bGh pAn has<br />
been shown to be as efficient as SV40 and HSV1tk polyadenylation signals in<br />
many different cell types 5 . The use of bGH pAn minimizes interference and<br />
possible recombination events with the SV40 polyadenylation signal. The pause<br />
site prevents transcriptional interference and read-through.<br />
R e f e re n c e s<br />
1- Kim et al (1990). Gene 2: 217-223.<br />
2- Takebe et al (1988). Mol. Cell Biol. 1: 466-472.<br />
3- Carswell et al(1989). Mol. Cell Biol. 10: 4248-4258.<br />
4- Levitt et al. (1989). Genes Dev. 7: 1019-1025.<br />
5- Goodwin et al. (1992). J. Biol. Chem. 23: 16330-16334.<br />
M E T H O D S<br />
Plasmid re s u s p e n s i o n :<br />
Quickly spin the tube containing the lyophilized plasmid to pellet the DNA. To<br />
obtain a plasmid solution at 1µg/µl, resuspend the DNA in 20µl of sterile H 2 O .<br />
Store resuspended plasmid at -20°C.<br />
Selection of bacteria with E. coli Fast-Media Blas:<br />
E. coli F a s t - M e d i a ® Blas is a n e w, fast and convenient way to prepare liquid and<br />
solid media for bacterial culture by using only a microwave. E. coli F a s t - M e d i a ®<br />
Blas is a TB (liquid) or LB (solid) based medium with blasticidin, and contains<br />
stabilizers. E. coli F a s t - M e d i a ® Blas can be ordered separately (catalog code # fasbl-l,<br />
fas-bl-s).<br />
M e t h o d :<br />
1- Pour the contents of a pouch into a clean borosilicate glass bottle or flask.<br />
2- Add 200 ml of distilled water to the flask<br />
3- Heat in a microwave on MEDIUM power setting (about 400Watts), until<br />
bubbles start appearing (approximately 3 minutes). Do not heat a closed<br />
c o n t a i n e r. Do not autoclave Fast-Media ® .<br />
4- Swirl gently to mix the preparation. Be careful, the bottle and media are hot,<br />
use heatproof pads or gloves and care when handling.<br />
5- Reheat the media for 30 seconds and gently swirl again. Repeat as necessary<br />
to completely dissolve the powder into solution. But be careful to avoid<br />
overboiling and volume loss.<br />
6- Let agar medium cool to 45˚C before pouring plates. Let liquid media cool to<br />
37˚C before seeding bacteria.<br />
N o t e : Do not reheat solidified Fast-Media ® as the antibiotic will be permanently<br />
d e s t royed by the pro c e d u re .<br />
TECHNICAL SUPPORT<br />
Toll free (US): 888-457-5873<br />
Outside US: (+1) 858-457-5873<br />
E-mail: info@invivogen.com<br />
Website: www.invivogen.com<br />
3950 Sorrento Valley Blvd. Suite A<br />
San Diego, CA 92121 - USA
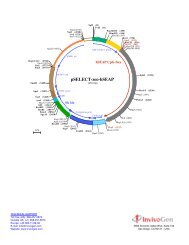
NotI (2952)<br />
XhoI (2947)<br />
SgfI (7)<br />
AgeI (90)<br />
PvuII (240)<br />
HindIII (246)<br />
EcoRI (2666)<br />
hEF1/HTLV prom<br />
EcoRV (551)<br />
SalI (630)<br />
BspHI (2237)<br />
Bsr<br />
EM7<br />
<strong>pBLAST42</strong>-<strong>mcs</strong><br />
(3116 bp)<br />
I117<br />
hIL2ss<br />
PvuII (659)<br />
SgrAI (669)<br />
EcoRI (743)<br />
NruI (747)<br />
AscI (753)<br />
BssHII (754)<br />
NheI (761)<br />
AseI (2180)<br />
HpaI (924)<br />
AvrII (2110)<br />
SV40 prom<br />
SwaI (1022)<br />
NcoI (2017)<br />
pMB1 Ori<br />
100
1<br />
101<br />
201<br />
301<br />
401<br />
501<br />
601<br />
701<br />
4<br />
801<br />
901<br />
1001<br />
1101<br />
1201<br />
1301<br />
1401<br />
1501<br />
1601<br />
1701<br />
1801<br />
1901<br />
2001<br />
2101<br />
SgfI (7) AgeI (90)<br />
GGATCTGCGATCGCTCCGGTGCCCGTCAGTGGGCAGAGCGCACATCGCCCACAGTCCCCGAGAAGTTGGGGGGAGGGGTCGGCAATTGAACCGGTGCCTA<br />
GAGAAGGTGGCGCGGGGTAAACTGGGAAAGTGATGTCGTGTACTGGCTCCGCCTTTTTCCCGAGGGTGGGGGAGAACCGTATATAAGTGCAGTAGTCGCC<br />
HindIII (246)<br />
PvuII (240)<br />
GTGAACGTTCTTTTTCGCAACGGGTTTGCCGCCAGAACACAGCTGAAGCTTCGAGGGGCTCGCATCTCTCCTTCACGCGCCCGCCGCCCTACCTGAGGCC<br />
GCCATCCACGCCGGTTGAGTCGCGTTCTGCCGCCTCCCGCCTGTGGTGCCTCCTGAACTGCGTCCGCCGTCTAGGTAAGTTTAAAGCTCAGGTCGAGACC<br />
GGGCCTTTGTCCGGCGCTCCCTTGGAGCCTACCTAGACTCAGCCGGCTCTCCACGCTTTGCCTGACCCTGCTTGCTCAACTCTACGTCTTTGTTTCGTTT<br />
EcoRV (551)<br />
TCTGTTCTGCGCCGTTACAGATCCAAGCTGTGACCGGCGCCTACgtaagtgatatctactagatttatcaaaaagagtgttgacttgtgagcgctcacaa<br />
SalI (630) PvuII (659) SgrAI (669)<br />
ttgatacttagattcatcgagagggacacgtcgactactaaccttcttctctttcctacagCTGAGATCACCGGCGAAGGAGGGCCACCATGTACAGGAT<br />
1 MetTyrArgMe<br />
BssHII (754)<br />
AscI (753)<br />
NruI (747)<br />
EcoRI (743)<br />
NheI (761)<br />
GCAACTCCTGTCTTGCATTGCACTAAGTCTTGCACTTGTCACGAATTCGCGAGGCGCGCCGCTAGCTCGACATGATAAGATACATTGATGAGTTTGGACA<br />
tGlnLeuLeuSerCysI leAlaLeuSerLeuAlaLeuValThrAsnSer<br />
AACCACAACTAGAATGCAGTGAAAAAAATGCTTTATTTGTGAAATTTGTGATGCTATTGCTTTATTTGTGAAATTTGTGATGCTATTGCTTTATTTGTAA<br />
HpaI (924)<br />
CCATTATAAGCTGCAATAAACAAGTTAACAACAACAATTGCATTCATTTTATGTTTCAGGTTCAGGGGGAGGTGTGGGAGGTTTTTTAAAGCAAGTAAAA<br />
SwaI (1022)<br />
CCTCTACAAATGTGGTAGATCATTTAAATGTTAATTAAGAACATGTGAGCAAAAGGCCAGCAAAAGGCCAGGAACCGTAAAAAGGCCGCGTTGCTGGCGT<br />
TTTTCCATAGGCTCCGCCCCCCTGACGAGCATCACAAAAATCGACGCTCAAGTCAGAGGTGGCGAAACCCGACAGGACTATAAAGATACCAGGCGTTTCC<br />
CCCTGGAAGCTCCCTCGTGCGCTCTCCTGTTCCGACCCTGCCGCTTACCGGATACCTGTCCGCCTTTCTCCCTTCGGGAAGCGTGGCGCTTTCTCAATGC<br />
TCACGCTGTAGGTATCTCAGTTCGGTGTAGGTCGTTCGCTCCAAGCTGGGCTGTGTGCACGAACCCCCCGTTCAGCCCGACCGCTGCGCCTTATCCGGTA<br />
ACTATCGTCTTGAGTCCAACCCGGTAAGACACGACTTATCGCCACTGGCAGCAGCCACTGGTAACAGGATTAGCAGAGCGAGGTATGTAGGCGGTGCTAC<br />
AGAGTTCTTGAAGTGGTGGCCTAACTACGGCTACACTAGAAGAACAGTATTTGGTATCTGCGCTCTGCTGAAGCCAGTTACCTTCGGAAAAAGAGTTGGT<br />
AGCTCTTGATCCGGCAAACAAACCACCGCTGGTAGCGGTGGTTTTTTTGTTTGCAAGCAGCAGATTACGCGCAGAAAAAAAGGATCTCAAGAAGATCCTT<br />
TGATCTTTTCTACGGGGTCTGACGCTCAGTGGAACGAAAACTCACGTTAAGGGATTTTGGTCATGGCTAGTTAATTAAGCTGTACACTGTGGAATGTGTG<br />
TCAGTTAGGGTGTGGAAAGTCCCCAGGCTCCCCAGCAGGCAGAAGTATGCAAAGCATGCATCTCAATTAGTCAGCAACCAGGTGTGGAAAGTCCCCAGGC<br />
TCCCCAGCAGGCAGAAGTATGCAAAGCATGCATCTCAATTAGTCAGCAACCATAGTCCCGCCCCTAACTCCGCCCATCCCGCCCCTAACTCCGCCCAGTT<br />
NcoI (2017)<br />
CCGCCCATTCTCCGCCCCATGGCTGACTAATTTTTTTTATTTATGCAGAGGCCGAGGCCGCCTCTGCCTCTGAGCTATTCCAGAAGTAGTGAGGAGGCTT<br />
AvrII (2110) AseI (2180)<br />
TTTTGGAGGCCTAGGCTTTTGCAAAAAGCTCCCGGGAGCTTGTATATCCATTTTCGGATCTGATcagCACGTGTTGACAATTAATCATCGGCATAGTATA<br />
BspHI (2237)<br />
2201 TCGGCATAGTATAATACGACAAGGTGAGGAACTAAATCATGAAGACCTTCAACATCTCTCAGCAGGATCTGGAGCTGGTGGAGGTCGCCACTGAGAAGAT<br />
1 MetLysThrPheAsnI leSerGlnGlnAspLeuGluLeuValGluValAlaThrGluLysI l<br />
2301 CACCATGCTCTATGAGGACAACAAGCACCATGTCGGGGCGGCCATCAGGACCAAGACTGGGGAGATCATCTCTGCTGTCCACATTGAGGCCTACATTGGC<br />
21 eThrMetLeuTyrGluAspAsnLysHisHisValGlyAlaAlaI leArgThrLysThrGlyGluI leI leSerAlaValHisI leGluAlaTyrI leGly<br />
2401 AGGGTCACTGTCTGTGCTGAAGCCATTGCCATTGGGTCTGCTGTGAGCAACGGGCAGAAGGACTTTGACACCATTGTGGCTGTCAGGCACCCCTACTCTG<br />
55 ArgValThrValCysAlaGluAlaI leAlaI leGlySerAlaValSerAsnGlyGlnLysAspPheAspThrI leValAlaValArgHisProTyrSerA<br />
2501 ATGAGGTGGACAGATCCATCAGGGTGGTCAGCCCCTGTGGCATGTGCAGAGAGCTCATCTCTGACTATGCTCCTGACTGCTTTGTGCTCATTGAGATGAA<br />
88 spGluValAspArgSerI leArgValValSerProCysGlyMetCysArgGluLeuI leSerAspTyrAlaProAspCysPheValLeuI leGluMetAs<br />
EcoRI (2666)<br />
2601 TGGCAAGCTGGTCAAAACCACCATTGAGGAACTCATCCCCCTCAAGTACACCAGGAACTAAACCTGAATTCGCTAGAGGGCCCTATTCTATAGTGTCACC<br />
121 nGlyLysLeuValLysThrThrI leGluGluLeuI leProLeuLysTyrThrArgAsn•••<br />
2701 TAAATGCTAGAGCTCGCTGATCAGCCTCGACTGTGCCTTCTAGTTGCCAGCCATCTGTTGTTTGCCCCTCCCCCGTGCCTTCCTTGACCCTGGAAGGTGC<br />
2801 CACTCCCACTGTCCTTTCCTAATAAAATGAGGAAATTGCATCGCATTGTCTGAGTAGGTGTCATTCTATTCTGGGGGGTGGGGTGGGGCAGGACAGCAAG<br />
NotI (2952)<br />
XhoI (2947)<br />
2901 GGGGAGGATTGGGAAGACAATAGCAGGCATGCGCAGGGCCCAATTGCTCGAGCGGCCGCAATAAAATATCTTTATTTTCATTACATCTGTGTGTTGGTTT<br />
3001 TTTGTGTGAATCGTAACTAACATACGCTCTCCATCAAAACAAAACGAAACAAAACAAACTAGCAAAATAGGCTGTCCCCAGTGCAAGTGCAGGTGCCAGA<br />
3101 ACATTTCTCTATCGAA