Part 4
Part 4
Part 4
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
CAP area (mV*msec)<br />
0.1<br />
0.2<br />
0.3<br />
0.4<br />
0.5<br />
0.6<br />
0.7<br />
0.8<br />
0.9<br />
1.0<br />
1.1<br />
Clinical score<br />
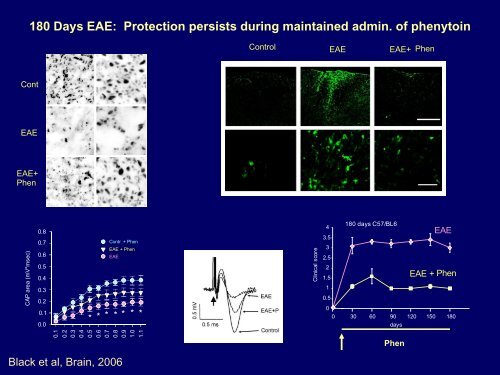
180 Days EAE: Protection persists during maintained admin. of phenytoin<br />
Control EAE EAE+ Phen<br />
Cont<br />
EAE<br />
EAE+<br />
Phen<br />
0.8<br />
0.7<br />
0.6<br />
Contr + Phen<br />
EAE + Phen<br />
EAE<br />
4<br />
3.5<br />
3<br />
2.5<br />
180 days C57/BL6<br />
EAE<br />
0.5<br />
0.4<br />
2<br />
1.5<br />
EAE + Phen<br />
0.3<br />
1<br />
0.2<br />
0.1<br />
0.0<br />
* * * * * * *<br />
0.5<br />
0<br />
0 30 60 90 120 150 180<br />
days<br />
Phen<br />
Black et al, Brain, 2006
Na V 1.6 Co-localizes with cytoskeletal F-actin, vimentin in TMP- human<br />
monocytic cell line, and in primary human macrophages<br />
shRNA knockdown of Na V 1.6, and Na channel block with TTX and<br />
phenytoin inhibit macrophage and microglial migration<br />
NaV1.6<br />
F-actin (phalloidin)<br />
Merge+DAPI<br />
Carrithers et al, J. Immunol. 2007<br />
Carrithers et al, JBC, 2008<br />
Microglia/6.3x10 5 m 2<br />
90<br />
80<br />
70<br />
60<br />
50<br />
40<br />
30<br />
20<br />
10<br />
0<br />
*<br />
#<br />
ACM ACM +<br />
ATP<br />
ACM +<br />
ATP +<br />
ACM +<br />
ATP +<br />
0.3 M TTX 10 M TTX<br />
#<br />
Black et al, Glia, 2009
I1848T and L858H Mutations Shift Activation,<br />
Slow Deactivation, and Enhance Response to Small, Slow<br />
Depolarizations to produce a Gain-of-Function in Na V 1.7<br />
0.0<br />
-0.2<br />
-0.4<br />
-0.6<br />
-0.8<br />
-1.0<br />
-80 -60 -40 -20 0 20 40<br />
test potential (mV)<br />
hNa v1.7<br />
l848T<br />
L858H<br />
-100<br />
4ms<br />
-20 -50<br />
hNa v 1.7<br />
l848T<br />
L858H<br />
percentage of peak current<br />
1<br />
0<br />
-1<br />
-2<br />
-3<br />
-100 -80 -60 -40 -20 0<br />
voltage (mV)<br />
hNav1.7<br />
I848T<br />
L858H<br />
0<br />
-100<br />
500 msec<br />
Cummins et al, J.Neurosci. 2004
Mutations in Exon 15 of SCN9A<br />
in Two Chinese families<br />
ATT Isoleucine (I 848)<br />
A CT<br />
Threonine (T)<br />
CTC Leucine (L 858)<br />
C AC<br />
Histidine (H)<br />
Promoter<br />
Exons<br />
Scn9A<br />
5N 5A<br />
Exon 15<br />
I II III IV<br />
7<br />
3 5<br />
15<br />
14 19 20 23 25 26<br />
NH 2<br />
2 4 6<br />
1<br />
Na1.7<br />
v<br />
8<br />
10<br />
9<br />
13<br />
L1<br />
12<br />
11<br />
16<br />
L2<br />
18<br />
17<br />
21<br />
22<br />
L3<br />
24<br />
COOH<br />
J. Med. Genet., March 2004
Molecular Pathophysiology of a Human<br />
Hereditary Pain Syndrome: Inherited Erythromelagia<br />
Sulayman Dib-Hajj, PhD<br />
Lynda Tyrrell, MSc<br />
Ted Cummins, PhD<br />
Tony Rush, PhD<br />
Angelika Lampert, MD<br />
Tanya Fischer, MD, PhD<br />
International Collaborators:<br />
Yong Yang, MD, PhD (China)<br />
Joost Drenth, MD (Netherlands)<br />
Bonnie Wallace, PhD (UK)<br />
Andreas O’Reilly, PhD (UK)<br />
Jin Choi, PhD<br />
Xiaoyang Cheng, PhD<br />
Mark Estacion, PhD<br />
Steve Waxman, MD, PhD<br />
ChongYang Han, PhD
T.M. Highly decorated police officer<br />
Gun shot wound<br />
• College graduate. Mult. Citations for bravery, community service.<br />
• 29 y.o.: GSW to arm. Injury to radial nerve. Distinguished Service<br />
Commendation.<br />
• Motor deficit, partial sensory loss in radial n. distribution.<br />
• Intermittent severe burning pain in area of sensory loss. Exacerbated by<br />
tactile and thermal stimuli.<br />
• No response to NSAIDs, tricyclics.<br />
• Minimal improvement w. phenytoin, gabapentin, lidocaine<br />
patch etc. Minimal response to TENS.<br />
• <strong>Part</strong>ial relief w/ carbamazepine, opiates. Doses limited by SEs.<br />
• Followed by neurologist, pain specialist, psychiatrist<br />
• Unable to work regularly. Disabled
Sodium Channels and Small Fiber Neuropathy
Na V 1.7 mutations in small fiber neuropathy<br />
Clinical:<br />
Janneke Hoeijmakers, MD<br />
Giuseppe Lauria, MD<br />
Joost Drenth, MD, PhD<br />
E.K. Van Houtte, MD<br />
Molecular Biology:<br />
Sulayman Dib-Hajj, PhD<br />
Lynda Tyrrell, MA<br />
Palak Shah, MA<br />
Larry Macala, MA<br />
Peng Zhao, PhD<br />
Cell Biology:<br />
Anna-Karin Persson, PhD<br />
Andreas Gasser, PhD<br />
Joel Black, PhD<br />
Physiology / Biophysics:<br />
Chongyang Han, PhD<br />
Xiaoyang Chen, PhD<br />
Hyesook Ahn, PhD<br />
Jin Choi, PhD<br />
Mark Estacion, PhD<br />
Catherina Faber, MD, PhD<br />
Ingemar Merkies, MD, PhD<br />
Steve Waxman, MD, PhD
Small Fiber Neuropathy<br />
• Dysfunction of small myelinated and unmyelinated nerve fibers:<br />
‣ Burning pain (feet > hands)<br />
‣ Autonomic sx (orthostatic hypotension,<br />
impotence, palpitations etc)<br />
‣ Dx: Normal neurol. exam (DTR’s, Vibr.<br />
sensation, Muscle Str.)<br />
‣ EMG, NCV: wnl<br />
A<br />
Patient<br />
‣ Dx confirmed by:<br />
Reduced IENFD on Cutan. Nerve Bx<br />
QST (warmth,cold sensation, heat pain)<br />
B<br />
Gender- and Age-matched Control
Can be associated with:<br />
Small Fiber Neuropathy<br />
Diabetes mellitus, IGT<br />
Hyperlipidemia, Hemochromatosis<br />
Liver, kidney, thyroid dysfn<br />
Amyloidosis<br />
Fabry’s disease<br />
Neurotoxic drugs (e.g. chemotx)<br />
Anti-phospholipid Ab syndrome<br />
Connective tissue disease<br />
Sarcoidosis<br />
Celiac disease<br />
HIV<br />
EtOH abuse<br />
Idiopathic (No Apparent Cause): 25% - 93% in diff. series.
SCN9A mutations: 8 of 28 pts with biopsyconfirmed<br />
SFN<br />
Patients referred with a<br />
clinical diagnosis of Small<br />
Fiber Neuropathy (SFN)<br />
N=248<br />
No underlying causes<br />
identified<br />
N=63 (25.4%)<br />
Underlying causes<br />
identified<br />
N=185 (74.6%)*<br />
Lost to follow-up or refused<br />
further participation<br />
N=19<br />
Excluded<br />
Patients with<br />
idiopathic SFN<br />
N=44<br />
IENFD + QST normal<br />
N=3<br />
IENFD or QST abnormal<br />
N=13<br />
IENFD + QST abnormal<br />
N=28<br />
Excluded<br />
No mutation<br />
found in SCN9A<br />
N=20<br />
Mutation found<br />
in SCN9A<br />
N=8<br />
Faber et al, Ann. Neurol., 2011
Nav1.7 mutations in Idiopathic SFN<br />
Domain I Domain II Domain III Domain IV<br />
M1532I<br />
+<br />
+<br />
+<br />
+<br />
+<br />
+<br />
+<br />
+<br />
+<br />
+<br />
+<br />
S1 S2 S3 S4 S5 S6 S1 S2 S3 S4 S5 S6 S1 S2 S3 S4 S5 S6 S1 S2 S3 S4 S5 S6<br />
+<br />
+<br />
+<br />
+<br />
+<br />
+<br />
+<br />
+<br />
+<br />
+<br />
extracellular<br />
cytoplasm<br />
N<br />
R185H*<br />
I228M<br />
L1 L2 L3<br />
I739V<br />
I720K<br />
D623N<br />
M932L – V991L<br />
C<br />
Faber et al, Ann. Neurol., 2011
R M P (m V )<br />
C u rre n t T h re sh o ld (p A )<br />
A P s /5 0 0 m s<br />
A1 A2 A3a<br />
WT<br />
1nA<br />
2ms<br />
M932L/V991L<br />
I /Im a x<br />
1.0<br />
0.8<br />
0.6<br />
0.4<br />
WT<br />
ML/VL<br />
1.0<br />
0.8<br />
0.6<br />
0.4<br />
G /G m a x<br />
I/ Im ax<br />
1.0<br />
0.8<br />
0.6<br />
0.4<br />
M932L / V991L<br />
WT<br />
ML/VL<br />
0.2<br />
0.0<br />
V 1/2,fast-inact<br />
-68.0±0.6mV<br />
-68.5±0.6mV<br />
V 1/2,act<br />
-20.2±0.6mV<br />
-20.4±1.2mV<br />
0.2<br />
0.0<br />
0.2<br />
0.0<br />
V 1/2,slow-inact<br />
-66.1±1.0mV<br />
-63.8±1.7mV<br />
-120 -80 -40 0 40<br />
Voltage (mV)<br />
-140 -120 -100 -80 -60 -40 -20 0 20<br />
Voltage (mV)<br />
A3b<br />
A4<br />
-35<br />
WT<br />
200pA<br />
20ms<br />
1.79% 1/11 (9.1%) 5.111.22% ± 5/10 (50%)<br />
A5<br />
ML/VL<br />
*<br />
A6<br />
4<br />
3<br />
2<br />
1<br />
0<br />
*<br />
WT<br />
ML/VL<br />
*<br />
*<br />
*<br />
*<br />
*<br />
*<br />
*<br />
*<br />
*<br />
*<br />
*<br />
*<br />
250<br />
-40<br />
200<br />
0 50 100 150 200 250 300 350 400 450 500<br />
Stimulus (pA)<br />
-45<br />
-50<br />
150<br />
**<br />
A7<br />
2.0 ± 0.3 Hz<br />
-55<br />
**<br />
100<br />
50<br />
4/2715; %<br />
(0/20; 0%) ; %<br />
-60<br />
WT<br />
ML/VL<br />
0<br />
WT<br />
ML/VL<br />
=+15% (p=0.072)
‘ When Hodgkin and I finished writing the 1952<br />
papers, each of us moved to other lines of work<br />
because we could not see how to make progress on<br />
the mechanism of channel action. Any idea of<br />
analyzing the channels by patch clamp or molecular<br />
biology would have seemed to us to be… science<br />
fiction ‘.<br />
Andrew Huxley, in The Axon, 1995
Na V 1.5 channels, localized on the endosomal membrane,<br />
provide a route for Na efflux that offsets proton influx during<br />
acidification of phagocytosed material<br />
Activation of Na V 1.5 with Veratridine triggers Na efflux and proton influx<br />
(acidification) of purified THP-1 endosomes<br />
Carrithers et al J. Immunol., 2007
Membrane potential (mV)<br />
E Na<br />
+40<br />
+20<br />
-20<br />
V<br />
30<br />
g Na<br />
20<br />
g K<br />
mS/cm 2<br />
E K<br />
-40<br />
-60<br />
-80<br />
10<br />
0<br />
4 msec<br />
I<br />
g<br />
Na<br />
m<br />
3<br />
h(<br />
V<br />
E<br />
Na<br />
)<br />
g<br />
K<br />
n<br />
4<br />
( V<br />
E<br />
K<br />
)<br />
g<br />
leak<br />
( V<br />
E<br />
leak<br />
)<br />
Hodgkin and Huxley, 1952
EFNS guidelines on skin biopsy 2010<br />
Skin biopsy: reduced IENFD<br />
patient<br />
control<br />
A<br />
B<br />
• 3 sections (50 μm) immunostained with PGP 9.5