tumor cell biology program - Sylvester Comprehensive Cancer Center
tumor cell biology program - Sylvester Comprehensive Cancer Center
tumor cell biology program - Sylvester Comprehensive Cancer Center
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
sis, and proceeds in a manner that has<br />
many similarities to eukaryotic transcription<br />
initiation.<br />
Dr. Malhotra’s laboratory is studying<br />
the mechanism of sigma54-enhancer<br />
directed transcription initiation using X-<br />
ray crystallography. Crystallization work<br />
on sigma54 and an associated activator<br />
protein is in progress. His team is also<br />
looking at the overall architecture of the<br />
sigma54-core RNAP activated complexes<br />
by 3D electron microscopy, and biochemical<br />
approaches such as protein-protein<br />
footprinting and cross-linking.<br />
Negative strand RNA viruses (involved<br />
in diseases such as measles, rabies<br />
and ebola) encode a multifunctional<br />
RNA dependent RNA polymerase,<br />
which performs both transcriptive and<br />
replicative functions and also displays<br />
mRNA capping and polyadenylation<br />
activities. Crystallographic studies of this<br />
polymerase and associated proteins are<br />
in progress in his laboratory.<br />
Ribosomal Structure<br />
Ribosomes are complex ribonucleoprotein<br />
particles responsible for mRNA directed<br />
protein synthesis in all living <strong>cell</strong>s.<br />
The large size of this macromolecular<br />
assembly (the E. coli ribosome has a MW<br />
of about 2.7 million daltons) makes it<br />
necessary to use both electron microscopy<br />
and X-ray crystallography to understand<br />
its structure. His laboratory is<br />
using 3-D electron-microscopy data<br />
from the laboratory of Dr. Joachim<br />
Frank, a Howard Hughes Research Investigator<br />
at the Wadsworth laboratories<br />
in Albany, New York, to understand the<br />
overall architecture of the ribosomal<br />
RNA in the ribosome. In addition, the<br />
structure of some ribosomal proteins and<br />
protein-RNA complexes by X-ray crystallography<br />
is also being investigated.<br />
Structural Studies of Bacterial<br />
Exoribonuclease<br />
Ribonucleases (RNases) play a central<br />
role in several aspects of <strong>cell</strong>ular RNA<br />
metabolism, including mRNA decay<br />
and maturation and turnover of structural<br />
RNAs. In collaboration with the<br />
laboratory of Dr. Murray Deutscher in<br />
the Department of Biochemistry and<br />
Molecular Biology, crystallographic studies<br />
are underway to decipher the atomic<br />
structure of RNase T from Escherichia<br />
coli.<br />
PUBLICATIONS<br />
Malhotra, A and Frank, J. Predicting<br />
three-dimensional structure of ribosomal<br />
RNA using cryo-electron<br />
microscopy maps of the E. coli ribosome.<br />
Nucleic Acid Research Symposium Series<br />
41: 32, 1999.<br />
Akila Mayeda, Ph.D.<br />
Assistant Professor of Biochemistry<br />
and Molecular Biology<br />
DESCRIPTION OF RESEARCH<br />
Dr. Mayeda’s research focuses on the<br />
molecular mechanisms of constitutive<br />
and alternative pre-mRNA splicing.<br />
By taking advantage of biochemical,<br />
molecular-biological, and immunological<br />
approaches together with splicing<br />
assays in vitro and in vivo, Dr. Mayeda’s<br />
team is attempting to purify and clone<br />
novel splicing factors that are involved<br />
in the general splicing pathways and in<br />
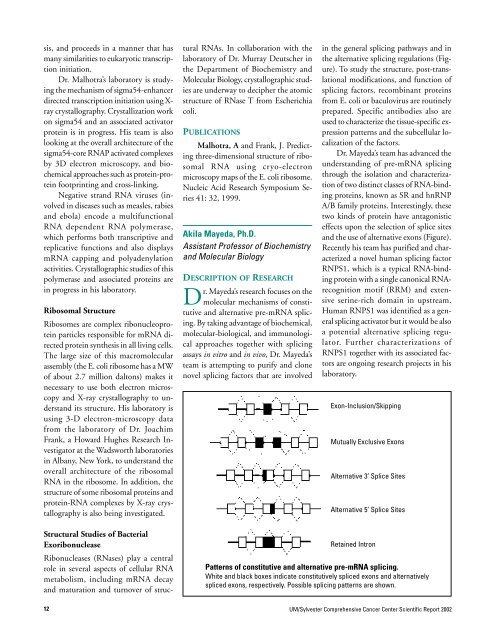
the alternative splicing regulations (Figure).<br />
To study the structure, post-translational<br />
modifications, and function of<br />
splicing factors, recombinant proteins<br />
from E. coli or baculovirus are routinely<br />
prepared. Specific antibodies also are<br />
used to characterize the tissue-specific expression<br />
patterns and the sub<strong>cell</strong>ular localization<br />
of the factors.<br />
Dr. Mayeda’s team has advanced the<br />
understanding of pre-mRNA splicing<br />
through the isolation and characterization<br />
of two distinct classes of RNA-binding<br />
proteins, known as SR and hnRNP<br />
A/B family proteins. Interestingly, these<br />
two kinds of protein have antagonistic<br />
effects upon the selection of splice sites<br />
and the use of alternative exons (Figure).<br />
Recently his team has purified and characterized<br />
a novel human splicing factor<br />
RNPS1, which is a typical RNA-binding<br />
protein with a single canonical RNArecognition<br />
motif (RRM) and extensive<br />
serine-rich domain in upstream.<br />
Human RNPS1 was identified as a general<br />
splicing activator but it would be also<br />
a potential alternative splicing regulator.<br />
Further characterizations of<br />
RNPS1 together with its associated factors<br />
are ongoing research projects in his<br />
laboratory.<br />
Exon-Inclusion/Skipping<br />
Mutually Exclusive Exons<br />
Alternative 3’ Splice Sites<br />
Alternative 5’ Splice Sites<br />
Retained Intron<br />
Patterns of constitutive and alternative pre-mRNA splicing.<br />
White and black boxes indicate constitutively spliced exons and alternatively<br />
spliced exons, respectively. Possible splicing patterns are shown.<br />
12<br />
UM/<strong>Sylvester</strong> <strong>Comprehensive</strong> <strong>Cancer</strong> <strong>Center</strong> Scientific Report 2002