Quantitative effects of composting state variables on ... - ResearchGate
Quantitative effects of composting state variables on ... - ResearchGate
Quantitative effects of composting state variables on ... - ResearchGate
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
1248 W. Sun et al. / Science <str<strong>on</strong>g>of</str<strong>on</strong>g> the Total Envir<strong>on</strong>ment 409 (2011) 1243–1254<br />
Best, Worst, and Mean Scores<br />
Average Distance<br />
Times Calling Proxy Table<br />
0.9<br />
0.8<br />
0.7<br />
0.6<br />
0.5<br />
0.4<br />
0.3<br />
0.2<br />
0.1<br />
3.5<br />
3<br />
2.5<br />
2<br />
1.5<br />
1<br />
0.5<br />
0<br />
100<br />
90<br />
80<br />
70<br />
60<br />
50<br />
40<br />
30<br />
20<br />
10<br />
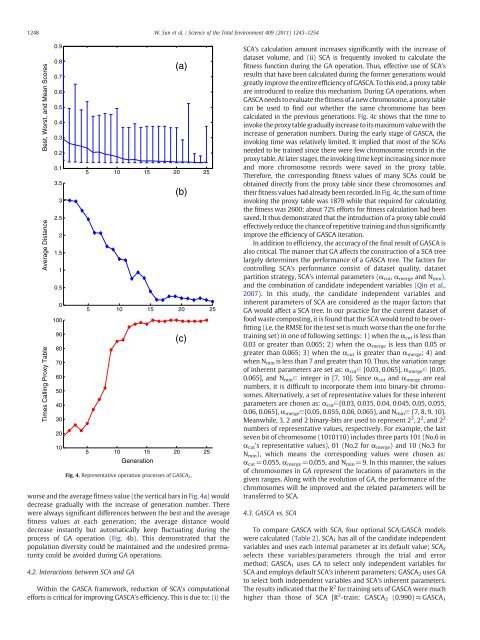
worse and the average fitness value (the vertical bars in Fig. 4a) would<br />
decrease gradually with the increase <str<strong>on</strong>g>of</str<strong>on</strong>g> generati<strong>on</strong> number. There<br />
were always significant differences between the best and the average<br />
fitness values at each generati<strong>on</strong>; the average distance would<br />
decrease instantly but automatically keep fluctuating during the<br />
process <str<strong>on</strong>g>of</str<strong>on</strong>g> GA operati<strong>on</strong> (Fig. 4b). This dem<strong>on</strong>strated that the<br />
populati<strong>on</strong> diversity could be maintained and the undesired prematurity<br />
could be avoided during GA operati<strong>on</strong>s.<br />
4.2. Interacti<strong>on</strong>s between SCA and GA<br />
5 10 15 20 25<br />
5 10 15 20 25<br />
5 10 15 20 25<br />
Generati<strong>on</strong><br />
(a)<br />
(b)<br />
(c)<br />
Fig. 4. Representative operati<strong>on</strong> processes <str<strong>on</strong>g>of</str<strong>on</strong>g> GASCA 2 .<br />
Within the GASCA framework, reducti<strong>on</strong> <str<strong>on</strong>g>of</str<strong>on</strong>g> SCA's computati<strong>on</strong>al<br />
efforts is critical for improving GASCA's efficiency. This is due to: (i) the<br />
SCA's calculati<strong>on</strong> amount increases significantly with the increase <str<strong>on</strong>g>of</str<strong>on</strong>g><br />
dataset volume, and (ii) SCA is frequently invoked to calculate the<br />
fitness functi<strong>on</strong> during the GA operati<strong>on</strong>. Thus, effective use <str<strong>on</strong>g>of</str<strong>on</strong>g> SCA's<br />
results that have been calculated during the former generati<strong>on</strong>s would<br />
greatly improve the entire efficiency <str<strong>on</strong>g>of</str<strong>on</strong>g> GASCA. To this end, a proxy table<br />
are introduced to realize this mechanism. During GA operati<strong>on</strong>s, when<br />
GASCA needs to evaluate the fitness <str<strong>on</strong>g>of</str<strong>on</strong>g> a new chromosome, a proxy table<br />
can be used to find out whether the same chromosome has been<br />
calculated in the previous generati<strong>on</strong>s. Fig. 4c shows that the time to<br />
invoke the proxy table gradually increase to its maximum value with the<br />
increase <str<strong>on</strong>g>of</str<strong>on</strong>g> generati<strong>on</strong> numbers. During the early stage <str<strong>on</strong>g>of</str<strong>on</strong>g> GASCA, the<br />
invoking time was relatively limited. It implied that most <str<strong>on</strong>g>of</str<strong>on</strong>g> the SCAs<br />
needed to be trained since there were few chromosome records in the<br />
proxy table. At later stages, the invoking time kept increasing since more<br />
and more chromosome records were saved in the proxy table.<br />
Therefore, the corresp<strong>on</strong>ding fitness values <str<strong>on</strong>g>of</str<strong>on</strong>g> many SCAs could be<br />
obtained directly from the proxy table since these chromosomes and<br />
their fitness values had already been recorded. In Fig. 4c, the sum <str<strong>on</strong>g>of</str<strong>on</strong>g> time<br />
invoking the proxy table was 1879 while that required for calculating<br />
the fitness was 2600; about 72% efforts for fitness calculati<strong>on</strong> had been<br />
saved. It thus dem<strong>on</strong>strated that the introducti<strong>on</strong> <str<strong>on</strong>g>of</str<strong>on</strong>g> a proxy table could<br />
effectively reduce the chance <str<strong>on</strong>g>of</str<strong>on</strong>g> repetitive training and thus significantly<br />
improve the efficiency <str<strong>on</strong>g>of</str<strong>on</strong>g> GASCA iterati<strong>on</strong>.<br />
In additi<strong>on</strong> to efficiency, the accuracy <str<strong>on</strong>g>of</str<strong>on</strong>g> the final result <str<strong>on</strong>g>of</str<strong>on</strong>g> GASCA is<br />
also critical. The manner that GA affects the c<strong>on</strong>structi<strong>on</strong> <str<strong>on</strong>g>of</str<strong>on</strong>g> a SCA tree<br />
largely determines the performance <str<strong>on</strong>g>of</str<strong>on</strong>g> a GASCA tree. The factors for<br />
c<strong>on</strong>trolling SCA's performance c<strong>on</strong>sist <str<strong>on</strong>g>of</str<strong>on</strong>g> dataset quality, dataset<br />
partiti<strong>on</strong> strategy, SCA's internal parameters (α cut , α merge and N min ),<br />
and the combinati<strong>on</strong> <str<strong>on</strong>g>of</str<strong>on</strong>g> candidate independent <str<strong>on</strong>g>variables</str<strong>on</strong>g> (Qin et al.,<br />
2007). In this study, the candidate independent <str<strong>on</strong>g>variables</str<strong>on</strong>g> and<br />
inherent parameters <str<strong>on</strong>g>of</str<strong>on</strong>g> SCA are c<strong>on</strong>sidered as the major factors that<br />
GA would affect a SCA tree. In our practice for the current dataset <str<strong>on</strong>g>of</str<strong>on</strong>g><br />
food waste <str<strong>on</strong>g>composting</str<strong>on</strong>g>, it is found that the SCA would tend to be overfitting<br />
(i.e. the RMSE for the test set is much worse than the <strong>on</strong>e for the<br />
training set) in <strong>on</strong>e <str<strong>on</strong>g>of</str<strong>on</strong>g> following settings: 1) when the α cut is less than<br />
0.03 or greater than 0.065; 2) when the α merge is less than 0.05 or<br />
greater than 0.065; 3) when the α cut is greater than α merge ; 4) and<br />
when N min is less than 7 and greater than 10. Thus, the variati<strong>on</strong> range<br />
<str<strong>on</strong>g>of</str<strong>on</strong>g> inherent parameters are set as: α cut ∈ [0.03, 0.065], α merge ∈ [0.05,<br />
0.065], and N min ∈ integer in [7, 10]. Since α cut and α merge are real<br />
numbers, it is difficult to incorporate them into binary-bit chromosomes.<br />
Alternatively, a set <str<strong>on</strong>g>of</str<strong>on</strong>g> representative values for these inherent<br />
parameters are chosen as: α cut ∈{0.03, 0.035, 0.04, 0.045, 0.05, 0.055,<br />
0.06, 0.065}, α merge ∈{0.05, 0.055, 0.06, 0.065}, and N min ∈ {7, 8, 9, 10}.<br />
Meanwhile, 3, 2 and 2 binary-bits are used to represent 2 3 ,2 2 , and 2 2<br />
numbers <str<strong>on</strong>g>of</str<strong>on</strong>g> representative values, respectively. For example, the last<br />
seven bit <str<strong>on</strong>g>of</str<strong>on</strong>g> chromosome (1010110) includes three parts 101 (No.6 in<br />
α cut 's representative values), 01 (No.2 for α merge ) and 10 (No.3 for<br />
N min ), which means the corresp<strong>on</strong>ding values were chosen as:<br />
α cut =0.055, α merge =0.055, and N min =9. In this manner, the values<br />
<str<strong>on</strong>g>of</str<strong>on</strong>g> chromosomes in GA represent the locati<strong>on</strong>s <str<strong>on</strong>g>of</str<strong>on</strong>g> parameters in the<br />
given ranges. Al<strong>on</strong>g with the evoluti<strong>on</strong> <str<strong>on</strong>g>of</str<strong>on</strong>g> GA, the performance <str<strong>on</strong>g>of</str<strong>on</strong>g> the<br />
chromosomes will be improved and the related parameters will be<br />
transferred to SCA.<br />
4.3. GASCA vs. SCA<br />
To compare GASCA with SCA, four opti<strong>on</strong>al SCA/GASCA models<br />
were calculated (Table 2). SCA 1 has all <str<strong>on</strong>g>of</str<strong>on</strong>g> the candidate independent<br />
<str<strong>on</strong>g>variables</str<strong>on</strong>g> and uses each internal parameter at its default value; SCA 2<br />
selects these <str<strong>on</strong>g>variables</str<strong>on</strong>g>/parameters through the trial and error<br />
method; GASCA 1 uses GA to select <strong>on</strong>ly independent <str<strong>on</strong>g>variables</str<strong>on</strong>g> for<br />
SCA and employs default SCA's inherent parameters; GASCA 2 uses GA<br />
to select both independent <str<strong>on</strong>g>variables</str<strong>on</strong>g> and SCA's inherent parameters.<br />
The results indicated that the R 2 for training sets <str<strong>on</strong>g>of</str<strong>on</strong>g> GASCA were much<br />
higher than those <str<strong>on</strong>g>of</str<strong>on</strong>g> SCA [R 2 -train: GASCA 2 (0.990)≈ GASCA 1