Evolutionary Computation : A Unified Approach
Evolutionary Computation : A Unified Approach
Evolutionary Computation : A Unified Approach
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
6.4. REPRODUCTION-ONLY MODELS 157<br />
16<br />
14<br />
12<br />
Blend recombination (0.5) and 1/10 Gaussian mutation (2.0)<br />
Blend recombination (0.5) and 2/10 Gaussian mutation (2.0)<br />
Blend recombination (0.5) and 5/10 Gaussian mutation (2.0)<br />
Blend recombination (0.5) and 10/10 Gaussian mutation (2.0)<br />
Population Dispersion<br />
10<br />
8<br />
6<br />
4<br />
2<br />
0<br />
0 20 40 60 80 100<br />
Generations<br />
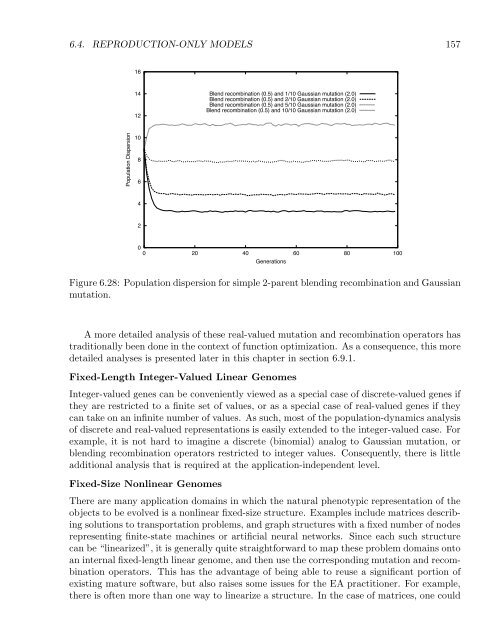
Figure 6.28: Population dispersion for simple 2-parent blending recombination and Gaussian<br />
mutation.<br />
A more detailed analysis of these real-valued mutation and recombination operators has<br />
traditionally been done in the context of function optimization. As a consequence, this more<br />
detailed analyses is presented later in this chapter in section 6.9.1.<br />
Fixed-Length Integer-Valued Linear Genomes<br />
Integer-valued genes can be conveniently viewed as a special case of discrete-valued genes if<br />
they are restricted to a finite set of values, or as a special case of real-valued genes if they<br />
can take on an infinite number of values. As such, most of the population-dynamics analysis<br />
of discrete and real-valued representations is easily extended to the integer-valued case. For<br />
example, it is not hard to imagine a discrete (binomial) analog to Gaussian mutation, or<br />
blending recombination operators restricted to integer values. Consequently, there is little<br />
additional analysis that is required at the application-independent level.<br />
Fixed-Size Nonlinear Genomes<br />
There are many application domains in which the natural phenotypic representation of the<br />
objects to be evolved is a nonlinear fixed-size structure. Examples include matrices describing<br />
solutions to transportation problems, and graph structures with a fixed number of nodes<br />
representing finite-state machines or artificial neural networks. Since each such structure<br />
can be “linearized”, it is generally quite straightforward to map these problem domains onto<br />
an internal fixed-length linear genome, and then use the corresponding mutation and recombination<br />
operators. This has the advantage of being able to reuse a significant portion of<br />
existing mature software, but also raises some issues for the EA practitioner. For example,<br />
there is often more than one way to linearize a structure. In the case of matrices, one could