tertiary structure of proteins involved in the bacillus subtilis cell ...
tertiary structure of proteins involved in the bacillus subtilis cell ...
tertiary structure of proteins involved in the bacillus subtilis cell ...
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
Materials Structure, vol. 10, number 1 (2003) 13<br />
TERTIARY STRUCTURE OF PROTEINS INVOLVED IN THE BACILLUS<br />
SUBTILIS CELL DIFFERENTIATION PROCESS<br />
Imrich Barák*, Katarína Muchová, Patrik Florek and Zuzana Chromiková<br />
In sti tute <strong>of</strong> Mo lec u lar Bi ol ogy, Slo vak Acad emy <strong>of</strong> Sci ences, Dúbravská cesta 21,<br />
845 51 Bratislava 45, Slovakia; phone: ++421 2 5930 7418, fax: ++421 2 5930 7416,<br />
e-mail: imrich.barak@savba.sk ,* Correspond<strong>in</strong>g author<br />
Keywords:<br />
crys tal log ra phy, pro te<strong>in</strong> struc ture, sporulation, Bacillus<br />
<strong>subtilis</strong><br />
Ab stract<br />
The num ber <strong>of</strong> pro te<strong>in</strong>s with func tions re stricted to spo -<br />
rulation <strong>in</strong> Bacillus <strong>subtilis</strong> is pro posed to be about 200.<br />
There is an enor mous amount <strong>of</strong> ge netic, bio chem i cal and<br />
molecular biology data about this simplest <strong>cell</strong> differentia -<br />
tion pro cess. How ever, at <strong>the</strong> pres ent, <strong>the</strong> num ber <strong>of</strong> known<br />
struc tures <strong>of</strong> sporulation spe cific pro te<strong>in</strong>s is about ten. This<br />
ar ti cle gives a short over view about re cent prog ress <strong>in</strong> crys -<br />
tal li za tion <strong>of</strong> key reg u la tors <strong>of</strong> sporulation.<br />
Ba cil lus <strong>subtilis</strong> as an ex am ple <strong>of</strong> pri mary<br />
<strong>cell</strong>ular differentiation<br />
Bacillus <strong>subtilis</strong> is a model or gan ism for <strong>the</strong> study <strong>of</strong> one <strong>of</strong><br />
<strong>the</strong> simplest <strong>cell</strong> differentiation processes, called sporulation.<br />
Thus this non patho genic soil bac te rium is <strong>of</strong> great<br />
im por tance for ba sic re search <strong>in</strong> gen eral and most likely it<br />
will be <strong>the</strong> first or gan ism <strong>in</strong> which <strong>the</strong> <strong>cell</strong> dif fer en ti a tion<br />
pro cess will be un der stood at <strong>the</strong> mo lec u lar level. The<br />
study <strong>of</strong> this pro cess has ad vanced to solv <strong>in</strong>g <strong>the</strong> struc ture<br />
<strong>of</strong> key pro te<strong>in</strong> reg u la tors. This re view fo cuses on <strong>the</strong> prog -<br />
ress <strong>in</strong> pro te<strong>in</strong> crys tal log ra phy ori ented to ward <strong>the</strong> un der -<br />
stand <strong>in</strong>g <strong>of</strong> sporulation mech a nisms, that have been<br />
achieved <strong>in</strong> <strong>the</strong> last cou ple <strong>of</strong> years. The life cy cle <strong>of</strong> B.<br />
<strong>subtilis</strong> comprises <strong>of</strong> three different processes: vegetative<br />
growth, sporulation and ger mi na tion - <strong>the</strong> spore out growth<br />
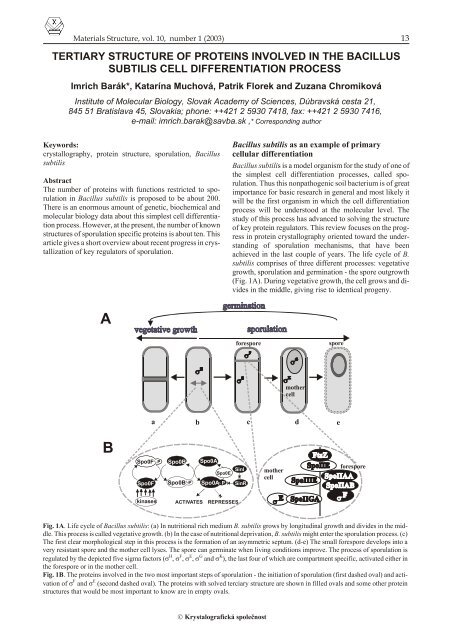
(Fig. 1A). Dur <strong>in</strong>g veg e ta tive growth, <strong>the</strong> <strong>cell</strong> grows and di -<br />
vides <strong>in</strong> <strong>the</strong> mid dle, giv <strong>in</strong>g rise to iden ti cal prog eny.<br />
A<br />
P<br />
forespore<br />
spore<br />
mo<strong>the</strong>r<br />
<strong>cell</strong><br />
a b c d<br />
e<br />
B<br />
Spo0F P<br />
Spo0F<br />
Spo0B<br />
Spo0B P<br />
Spo0A<br />
Spo0E<br />
Spo0A P<br />
S<strong>in</strong>I<br />
S<strong>in</strong>R<br />
mo<strong>the</strong>r<br />
<strong>cell</strong><br />
forespore<br />
k<strong>in</strong>ases<br />
ACTIVATES<br />
REPRESSES<br />
Fig. 1A. Life cy cle <strong>of</strong> Ba cil lus <strong>subtilis</strong>: (a) In nu tri tional rich me dium B. <strong>subtilis</strong> grows by lon gi tu di nal growth and di vides <strong>in</strong> <strong>the</strong> mid -<br />
dle. This pro cess is called veg e ta tive growth. (b) In <strong>the</strong> case <strong>of</strong> nu tri tional de pri va tion, B. <strong>subtilis</strong> might en ter <strong>the</strong> sporulation pro cess. (c)<br />
The first clear mor pho log i cal step <strong>in</strong> this pro cess is <strong>the</strong> for ma tion <strong>of</strong> an asym met ric sep tum. (d-e) The small forespore de vel ops <strong>in</strong>to a<br />
very re sis tant spore and <strong>the</strong> mo<strong>the</strong>r <strong>cell</strong> lyses. The spore can ger mi nate when liv <strong>in</strong>g con di tions im prove. The pro cess <strong>of</strong> sporulation is<br />
reg u lated by <strong>the</strong> de picted five sigma fac tors ( H , F , E , G and K ), <strong>the</strong> last four <strong>of</strong> which are com part ment spe cific, ac ti vated ei <strong>the</strong>r <strong>in</strong><br />
<strong>the</strong> forespore or <strong>in</strong> <strong>the</strong> mo<strong>the</strong>r <strong>cell</strong>.<br />
Fig. 1B. The pro te<strong>in</strong>s <strong>in</strong> volved <strong>in</strong> <strong>the</strong> two most im por tant steps <strong>of</strong> sporulation - <strong>the</strong> <strong>in</strong>i ti a tion <strong>of</strong> sporulation (first dashed oval) and ac ti -<br />
va tion <strong>of</strong> F and E (sec ond dashed oval). The pro te<strong>in</strong>s with solved terciary struc ture are shown <strong>in</strong> filled ovals and some o<strong>the</strong>r pro te<strong>in</strong><br />
struc tures that would be most im por tant to know are <strong>in</strong> empty ovals.<br />
Krystalografická spoleènost
14 TERTIARY STRUCTURE OF PROTEINS...<br />
A<br />
Spo0F<br />
Spo0B<br />
B<br />
Asp55~P<br />
N<br />
Thr84<br />
Phe103<br />
C<br />
N<br />
C<br />
áF<br />
áA<br />
áB<br />
áC<br />
áE<br />
áD<br />
C<br />
Spo0B<br />
Spo0F<br />
D<br />
E<br />
C<br />
F<br />
ó F<br />
S<strong>in</strong>R<br />
N<br />
SpoIIAB<br />
S<strong>in</strong>I<br />
Ser58~P<br />
SpoIIAB<br />
Fig. 2. Crys tal struc ture <strong>of</strong> some sporulation spe cific pro te<strong>in</strong>s. A. Com plex <strong>of</strong> two Spo0F mono mers with Spo0B dimer; am<strong>in</strong>oacid<br />
res i dues <strong>of</strong> Spo0F ac tive site are visu al ised on up per Spo0F mol e cule (PDB ID: 1F51). B. Ac tive site <strong>of</strong> N-Spo0A; phosphorylated<br />
Asp55 and am<strong>in</strong>oacid res i dues, which change <strong>the</strong>ir con for ma tion dur <strong>in</strong>g phosphorylation are shown (PDB ID: 1QMP). C. Com plex <strong>of</strong><br />
two C-ter mi nal do ma<strong>in</strong> <strong>of</strong> Spo0A <strong>in</strong> com plex with DNA; he li ces A-F are de picted (PDB ID: 1LQ1). D. Struc ture <strong>of</strong> S<strong>in</strong>R-S<strong>in</strong>I<br />
com plex (PDB ID: 1B0N). E. Phosphorylated form <strong>of</strong> SpoIIAA with site <strong>of</strong> phosphorylation po<strong>in</strong>t <strong>in</strong>g to sol vent (PDB ID: 1H4X). F.<br />
Com plex <strong>of</strong> SpoIIAB dimer with part <strong>of</strong> F mono mer (PDB ID: 1L0O).<br />
Ini ti a tion <strong>of</strong> sporulation<br />
Veg e ta tive <strong>cell</strong>s are able to mon i tor <strong>the</strong> num ber <strong>of</strong> <strong>cell</strong>s <strong>in</strong><br />
<strong>the</strong> pop u la tion and <strong>the</strong> amount <strong>of</strong> nu tri tional sub stances <strong>in</strong><br />
<strong>the</strong> en vi ron ment. In case <strong>of</strong> nu tri tion de pri va tion, <strong>the</strong> bac te -<br />
rial <strong>cell</strong> can en ter <strong>the</strong> pro cess <strong>of</strong> spore de vel op ment. There<br />
are many pro te<strong>in</strong>s <strong>in</strong> volved <strong>in</strong> <strong>the</strong> <strong>in</strong>i ti a tion stage <strong>of</strong><br />
sporulation that are im por tant for proper chro mo some seg -<br />
re ga tion, cytok<strong>in</strong>esis, <strong>cell</strong> length and <strong>cell</strong> shape de ter mi na -<br />
tion. Some <strong>of</strong> <strong>the</strong>se pro te<strong>in</strong>s play a cru cial role also dur <strong>in</strong>g<br />
sporulation. The <strong>in</strong>i ti a tion <strong>of</strong> sporulation de pends on a<br />
com plex se ries <strong>of</strong> ex ter nal and <strong>in</strong> ter nal sig nals and is str<strong>in</strong> -<br />
gently con trolled by a net work <strong>of</strong> reg u la tory pro te<strong>in</strong>s <strong>of</strong> <strong>the</strong><br />
phosphorelay. Re cently, <strong>the</strong> struc ture <strong>of</strong> some pro te<strong>in</strong>s <strong>in</strong> -<br />
volved <strong>in</strong> this adap tive re sponse were solved and ques tions<br />
partially answered regard<strong>in</strong>g <strong>in</strong>teractions between <strong>the</strong> compo<br />
nents <strong>of</strong> this reg u la tory net work.<br />
This phosphorelay is an ex panded ver sion <strong>of</strong> a twocom<br />
po nent sig nal transduction sys tem. The first com po -<br />
nent, a sen sor k<strong>in</strong>ase (up to five sporulation sen sor k<strong>in</strong> ases<br />
K<strong>in</strong> A-E have been iden ti fied [1]), is autophosphorylated<br />
by ATP on its histid<strong>in</strong>e res i due <strong>in</strong> re sponse to en vi ron men -<br />
tal changes. The phopshoryl group is sub se quently trans -<br />
ferred to <strong>the</strong> aspartyl res i due <strong>of</strong> <strong>the</strong> re sponse reg u la tor<br />
Spo0F. Then phosphotransferase Spo0B trans fers phos -<br />
phate from Spo0F to <strong>the</strong> fi nal re sponse reg u la tor,<br />
transcriptional ac ti va tor and repressor Spo0A [2]. Spo0F is<br />
a typ i cal s<strong>in</strong> gle do ma<strong>in</strong> re sponse reg u la tor that has an -<br />
struc ture with five -he li ces ar ranged around a cen tral<br />
-sheet con sist <strong>in</strong>g <strong>of</strong> five par al lel -strands [3] (Fig. 2A).<br />
The ac tive site com pris <strong>in</strong>g three aspartates Asp10, Asp11<br />
and Asp54, and <strong>the</strong> threon<strong>in</strong>e Thr82 and lys<strong>in</strong>e Lys104 res -<br />
i dues is sit u ated as a small pocket at <strong>the</strong> carboxy-ter mi nal<br />
end <strong>of</strong> <strong>the</strong> -sheet. The site <strong>of</strong> phosphorylation, Asp54 is at<br />
<strong>the</strong> bot tom <strong>of</strong> <strong>the</strong> pocket and is ac ces si ble to sol vent. The<br />
over all struc ture and ac tive site ge om e try <strong>of</strong> Spo0F is re -<br />
mark ably sim i lar to o<strong>the</strong>r reg u la tory do ma<strong>in</strong>s such as CheY<br />
[4], Fix J [5] or Spo0A [6].<br />
The phos phate ac cu mu lated on Spo0F is im me di ately<br />
de liv ered to Spo0A by <strong>the</strong> ac tion <strong>of</strong> Spo0B. Spo0B is a<br />
phosphotransferase with some func tional and struc tural<br />
sim i lar i ties with histid<strong>in</strong>e k<strong>in</strong> ases [7]. It is phosphorylated<br />
on its histid<strong>in</strong>e res i due and forms a dimer. The mono mer is<br />
made up <strong>of</strong> two do ma<strong>in</strong>s, an am<strong>in</strong>o-ter mi nal-helical hair -<br />
p<strong>in</strong> do ma<strong>in</strong> and a carboxy-ter mi nal do ma<strong>in</strong> with an /<br />
fold. The crys tal struc ture re veals that dimer is formed by<br />
<strong>the</strong> as so ci a tion <strong>of</strong> <strong>the</strong> hair p<strong>in</strong> he li cal do ma<strong>in</strong>s from two<br />
mol e cules to form a four-he lix bun dle with <strong>the</strong> site <strong>of</strong><br />
phosphorylation His30 pro trud <strong>in</strong>g <strong>in</strong>to <strong>the</strong> sol vent. There<br />
are two ac tive sites per dimer. The crys tal struc ture <strong>of</strong> <strong>the</strong><br />
com plex be tween Spo0F and Spo0B [8] re vealed how <strong>the</strong><br />
reg u la tory do ma<strong>in</strong> and phosphotransfer do ma<strong>in</strong> <strong>in</strong> ter act to -<br />
ge<strong>the</strong>r (Fig. 2A). The cocrystal con ta<strong>in</strong>ed two Spo0F mol e -<br />
cules per Spo0B dimer. The 1-he lix <strong>of</strong> each Spo0F<br />
mol e cule and <strong>the</strong> five - loops on <strong>the</strong> top <strong>of</strong> <strong>the</strong> mol e cule<br />
Krystalografická spoleènost
I. Barák, K. Muchová, P. Florek and Z. Chromiková 15<br />
<strong>in</strong>teract with <strong>the</strong> 1-he lix <strong>of</strong> Spo0B. This <strong>in</strong> ter ac tion aligns<br />
<strong>the</strong> histid<strong>in</strong>e <strong>of</strong> Spo0B with <strong>the</strong> aspartate <strong>of</strong> Spo0F <strong>in</strong> cor -<br />
rect con fig u ra tion and dis tance for phosphotransfer. The<br />
hy dro pho bic sur face <strong>of</strong> Spo0F com prised <strong>of</strong> hy dro pho bic<br />
res i dues, Ile15, Leu18, Pro105, Phe106 and Ile108, is <strong>in</strong> -<br />
volved <strong>in</strong> this <strong>in</strong> ter ac tion. S<strong>in</strong>ce <strong>the</strong> struc ture <strong>of</strong> Spo0B is<br />
very sim i lar to <strong>the</strong> struc ture <strong>of</strong> histid<strong>in</strong>e sen sor k<strong>in</strong> ases, <strong>the</strong><br />
Spo0B-Spo0F struc ture should be viewed as a model for<br />
re sponse reg u la tor-sen sor k<strong>in</strong>ase <strong>in</strong> ter ac tions and <strong>the</strong>se six<br />
highly con served res i dues may play a key role <strong>in</strong> this <strong>in</strong> ter -<br />
action.<br />
Spo0A mas ter con trol for en try <strong>in</strong>to sporulation<br />
The fi nal ac cep tor <strong>of</strong> phos phate mi gra tion via Spo0F and<br />
Spo0B is Spo0A, <strong>the</strong> re sponse reg u la tor and key con trol el -<br />
e ment <strong>in</strong> <strong>the</strong> de ci sion to sporulate [2]. Spo0A con sists <strong>of</strong><br />
two do ma<strong>in</strong>s, <strong>the</strong> N-ter mi nal phosphoacceptor (re ceiver)<br />
do ma<strong>in</strong> and <strong>the</strong> C-ter mi nal DNA-b<strong>in</strong>d <strong>in</strong>g (effector) do -<br />
ma<strong>in</strong> [9] con nected by a flex i ble l<strong>in</strong>ker pep tide. As <strong>the</strong><br />
DNA-b<strong>in</strong>d <strong>in</strong>g do ma<strong>in</strong> is ac tive af ter re moval <strong>of</strong> <strong>the</strong> N-ter -<br />
mi nal do ma<strong>in</strong>, this re ceiver do ma<strong>in</strong> is thought to <strong>in</strong> hibit <strong>the</strong><br />
effector do ma<strong>in</strong> and this auto<strong>in</strong>hibition is over come by<br />
conformational changes fol low <strong>in</strong>g phosphorylation <strong>of</strong> <strong>the</strong><br />
N-ter mi nal do ma<strong>in</strong>. The crys tal struc ture <strong>of</strong> N-Spo0A was<br />
de ter m<strong>in</strong>ed <strong>in</strong> both phosphorylated and unphosphorylated<br />
forms [6, 10]. The over all fold <strong>of</strong> N-Spo0A per fectly<br />
matches <strong>the</strong> struc tures <strong>of</strong> <strong>the</strong> reg u la tory do ma<strong>in</strong>s <strong>of</strong> o<strong>the</strong>r<br />
re sponse reg u la tors. The struc ture <strong>of</strong> N-Spo0A~P re vealed<br />
that phosphorylation <strong>of</strong> aspartate 55 leads to con -<br />
formational changes at <strong>the</strong> ac tive site (Fig. 2B). The re ori -<br />
en ta tion <strong>of</strong> <strong>the</strong> Thr84 hydroxyl group to wards <strong>the</strong><br />
phosphoryl group is ac com pa nied by move ment <strong>of</strong> a side<br />
cha<strong>in</strong> <strong>of</strong> Phe103 from a sol vent ex posed po si tion <strong>in</strong>to <strong>the</strong><br />
pro te<strong>in</strong> core. S<strong>in</strong>ce <strong>the</strong>se ac tive-site res i dues are highly<br />
con served among re sponse reg u la tors and sim i lar changes<br />
were ob served <strong>in</strong> <strong>the</strong> struc tures <strong>of</strong> o<strong>the</strong>r ac ti vated re sponse<br />
reg u la tors [5, 11], this "ar o matic switch" could rep re sent a<br />
gen eral mech a nism <strong>of</strong> sig nal prop a ga tion through <strong>the</strong> pro -<br />
te<strong>in</strong> fol low <strong>in</strong>g aspartate phosphorylation. More over,<br />
mutational and bio chem i cal stud ies sug gest that Phe103<br />
could be <strong>in</strong> volved <strong>in</strong> dimer for ma tion <strong>of</strong> Spo0A af ter<br />
phosphorylation (un pub lished data).<br />
Re cently, struc tures <strong>of</strong> <strong>the</strong> C-ter mi nal do ma<strong>in</strong> <strong>of</strong><br />
Spo0A [12] and C-Spo0A <strong>in</strong> com plex with DNA [13] also<br />
were solved. The C-Spo0A (Fig. 2C) forms a large he li cal<br />
as sem bly com pris <strong>in</strong>g <strong>of</strong> six -helices (A-F) connected by<br />
short pep tides. The struc ture con ta<strong>in</strong>s a clas si cal he lixturn-<br />
he lix (he li ces C and D) as a DNA b<strong>in</strong>d <strong>in</strong>g mo tif. He li -<br />
ces A, B and F form a three-he li cal bun dle lo cated atop <strong>the</strong><br />
HTH and he lix E pro trudes from <strong>the</strong> bun dle op po site <strong>the</strong><br />
HTH. The res i dues <strong>in</strong> volved <strong>in</strong> con tact with A sub unit <strong>of</strong><br />
RNA polymerase <strong>in</strong> <strong>the</strong> transcription <strong>in</strong>itiation complex are<br />
sit u ated at <strong>the</strong> op po site end <strong>of</strong> <strong>the</strong> DNA rec og ni tion he lix<br />
and form <strong>in</strong> con trast with <strong>the</strong> rest <strong>of</strong> <strong>the</strong> struc ture a flex i ble<br />
and mo bile seg ment. The crys tal struc ture <strong>of</strong> C-Spo0A <strong>in</strong><br />
com plex with its tar get DNA (Fig. 2C) re vealed that two<br />
mol e cules form a dimer upon b<strong>in</strong>d <strong>in</strong>g to <strong>the</strong> tan dem b<strong>in</strong>d -<br />
<strong>in</strong>g sites on <strong>the</strong> DNA. The mol e cules are bound <strong>in</strong> head to<br />
tail ar range ment such that <strong>the</strong> C-ter mi nus <strong>of</strong> one mol e cule<br />
(he lix F) <strong>in</strong> ter acts with <strong>the</strong> N-ter mi nus <strong>of</strong> an o<strong>the</strong>r (he lix B).<br />
He lix F <strong>of</strong> one mol e cule and he li ces A and B <strong>of</strong> <strong>the</strong> sec ond<br />
mol e cule form a C-Spo0A dimer <strong>in</strong> ter face. Such di me -<br />
rization should sta bi lize <strong>the</strong> pro te<strong>in</strong>-DNA com plex. Also<br />
analytical ultracentrifugation and gel filtration experiments<br />
re vealed dimerization <strong>of</strong> en tire Spo0A af ter phospho -<br />
rylation [14]. These re sults <strong>in</strong> di cate that dimerization af ter<br />
phosphorylation is me di ated pr<strong>in</strong> ci pally by <strong>the</strong> re ceiver do -<br />
ma<strong>in</strong> prob a bly fol lowed by dimerization <strong>of</strong> <strong>the</strong> effector do -<br />
ma<strong>in</strong> bound on DNA. Ob ta<strong>in</strong>ed struc tural and bio chem i cal<br />
data pro vide an im por tant frame work for fur <strong>the</strong>r <strong>in</strong> ter pre ta -<br />
tion <strong>of</strong> <strong>the</strong> mutational data <strong>of</strong> Spo0A and thus un der stand -<br />
<strong>in</strong>g <strong>of</strong> <strong>the</strong> bi o log i cal func tion <strong>of</strong> Spo0A at <strong>the</strong> mo lec u lar<br />
level.<br />
O<strong>the</strong>r pro te<strong>in</strong>s <strong>in</strong> volved <strong>in</strong> <strong>in</strong>i ti a tion <strong>of</strong><br />
sporulation<br />
To en sure that spore for ma tion is <strong>the</strong> only way which en -<br />
ables <strong>the</strong> bac te rium is able to over come un fa vour able en vi -<br />
ron men tal con di tions, <strong>the</strong> en try <strong>in</strong>to sporulation is<br />
pre cisely con trolled not only by <strong>the</strong> above men tioned com -<br />
po nents <strong>of</strong> phosphorelay, but also by o<strong>the</strong>r pro te<strong>in</strong>s.<br />
Among <strong>the</strong>se, an im por tant role be longs to <strong>the</strong> sporulation<br />
<strong>in</strong> hib i tor S<strong>in</strong>R and its an tag o nist S<strong>in</strong>I [15]. S<strong>in</strong>R is a<br />
tetrameric DNA b<strong>in</strong>d <strong>in</strong>g pro te<strong>in</strong> that con trols sporulation<br />
di rectly through re pres sion <strong>of</strong> spo0A and stage II spo -<br />
rulation genes, spoIIA, spoIIE and spoIIG. For spo rulation<br />
to pro ceed, <strong>the</strong> ac tiv ity <strong>of</strong> S<strong>in</strong>R must be switched <strong>of</strong>f. This<br />
is brought about by <strong>the</strong> ac tion <strong>of</strong> S<strong>in</strong>I which forms<br />
S<strong>in</strong>I-S<strong>in</strong>R com plex that is un able to b<strong>in</strong>d to DNA and<br />
<strong>the</strong>re fore <strong>the</strong> re pres sive ef fects <strong>of</strong> S<strong>in</strong>R on tran scrip tion are<br />
re lieved. The crys tal struc ture <strong>of</strong> this heterodimer [16] re -<br />
vealed an -he li cal as sem bly <strong>of</strong> two do ma<strong>in</strong>s <strong>of</strong> ap prox i -<br />
mately equal size, an oligomerisation do ma<strong>in</strong> and a<br />
DNA-b<strong>in</strong>d <strong>in</strong>g do ma<strong>in</strong>. The oligomerisation do ma<strong>in</strong> is<br />
formed by four -helices, two from <strong>the</strong> C-term<strong>in</strong>al residues<br />
<strong>of</strong> S<strong>in</strong>R and two from <strong>the</strong> cen tral res i dues <strong>of</strong> S<strong>in</strong>I (Fig. 2D).<br />
Re cently, struc ture <strong>of</strong> an o<strong>the</strong>r pro te<strong>in</strong>, Obg , a Spo0B-as so -<br />
ci ated GTP b<strong>in</strong>d <strong>in</strong>g pro te<strong>in</strong>, <strong>in</strong> volved <strong>in</strong> <strong>the</strong> sporulation<br />
path way, was solved [17]. This struc ture re vealed <strong>the</strong><br />
unique ar chi tec ture <strong>of</strong> a GTP b<strong>in</strong>d <strong>in</strong>g pro te<strong>in</strong> and to ge<strong>the</strong>r<br />
with biochemical analysis suggests a potential role for this<br />
pro te<strong>in</strong> <strong>in</strong> <strong>the</strong> life cy cle <strong>of</strong> B. <strong>subtilis</strong>.<br />
Pro te<strong>in</strong>s <strong>in</strong> volved <strong>in</strong> <strong>the</strong> ac ti va tion <strong>of</strong> first<br />
compartment specific sigma factor - F<br />
The first clear mor pho log i cal fea ture <strong>of</strong> sporulation is <strong>the</strong><br />
for ma tion <strong>of</strong> an asym met ric sep tum (Fig. 1A) that bi sects<br />
<strong>the</strong> bac te rial <strong>cell</strong> <strong>in</strong>to two un equally sized com part ments,<br />
<strong>the</strong> larger mo<strong>the</strong>r <strong>cell</strong> and <strong>the</strong> smaller forespore. The proper<br />
po si tion <strong>in</strong>g <strong>of</strong> this sporulation sep tum is de pend ent on<br />
Spo0A ac tiv ity, which me di ates <strong>the</strong> as sem bly <strong>of</strong> di vi sion<br />
pro te<strong>in</strong>s near <strong>the</strong> <strong>cell</strong> pole <strong>in</strong> stead <strong>of</strong> at mid-<strong>cell</strong>. Such po si -<br />
tional switch could be par tially trig gered through <strong>the</strong> ac tiv -<br />
ity <strong>of</strong> <strong>the</strong> sporulation spe cific pro te<strong>in</strong> SpoIIE, also a<br />
com po nent <strong>of</strong> <strong>the</strong> sporulation sep tum, that is ex pressed as a<br />
re sult <strong>of</strong> Spo0A ac tiv ity [18, 19, 20, 21]. The cen tral do -<br />
ma<strong>in</strong> <strong>of</strong> SpoIIE is <strong>in</strong> volved <strong>in</strong> oligomerization <strong>of</strong> <strong>the</strong> pro -<br />
te<strong>in</strong> and it is re spon si ble for <strong>in</strong> ter ac tion with FtsZ, <strong>the</strong><br />
pro te<strong>in</strong> es sen tial for <strong>cell</strong> di vi sion [22, 23]. SpoIIE pro te<strong>in</strong><br />
can dephosphorylate SpoIIAA-P and thus it plays a cru cial<br />
Krystalografická spoleènost
16 TERTIARY STRUCTURE OF PROTEINS...<br />
role <strong>in</strong> ac ti va tion <strong>of</strong> <strong>the</strong> first <strong>cell</strong>-type spe cific fac tor - F<br />
(Fig. 1B) [24, 25]. The crys tal struc ture <strong>of</strong> phosphorylated<br />
and unphosphorylated form <strong>of</strong> SpoIIAA was solved from<br />
Ba cil lus sphaericus [26]. This s<strong>in</strong> gle do ma<strong>in</strong> glob u lar pro -<br />
te<strong>in</strong> con sists <strong>of</strong> a cen tral sheet <strong>of</strong> five strands, and four<br />
he li ces (Fig. 2E). On one face, <strong>the</strong> -pleated sheet is em -<br />
bed ded with a pair <strong>of</strong> -he li ces, while <strong>the</strong> o<strong>the</strong>r face is ex -<br />
posed to <strong>the</strong> sol vent. The phosphorylation site,<br />
de pho sphorylation <strong>of</strong> which is im por tant for ac ti va tion <strong>of</strong><br />
F , is lo cated at <strong>the</strong> N-ter mi nus <strong>of</strong> <strong>the</strong> he lix 2 so that <strong>the</strong><br />
pho spho ryl group po<strong>in</strong>ts <strong>in</strong>to <strong>the</strong> sol vent. Com par i sons be -<br />
tween unphosphorylated and phosphorylated SpoIIAA had<br />
shown, that struc tural changes ac com pa ny <strong>in</strong>g phospho -<br />
rylation are only slight. Ex tended hy dro pho bic sur faces to -<br />
ge<strong>the</strong>r with flex i bil ity <strong>in</strong> he lix 3 and <strong>the</strong> fol low <strong>in</strong>g loop<br />
<strong>in</strong> di cate that this pro te<strong>in</strong> <strong>of</strong> ten un der goes a dis or der to or -<br />
der tran si tion and <strong>the</strong>re fore has a dis po si tion for form <strong>in</strong>g<br />
sta ble pro te<strong>in</strong>-pro te<strong>in</strong> <strong>in</strong> ter ac tions [26]. Also <strong>the</strong> struc ture<br />
<strong>of</strong> <strong>the</strong> anti- fac tor SpoIIAB <strong>in</strong> <strong>the</strong> com plex with F <strong>of</strong> Ba -<br />
cil lus stearo<strong>the</strong>rmophilus was re solved [27]. SpoIIAB - F<br />
com plex con sists <strong>of</strong> a SpoIIAB dimer bound to one F mol -<br />
e cule so that F b<strong>in</strong>d <strong>in</strong>g di rectly to <strong>the</strong> dimer <strong>in</strong> ter face<br />
makes con tact with both SpoIIAB mono mers (Fig. 2F).<br />
The whole struc ture <strong>of</strong> F is com prised <strong>of</strong> three com pactly<br />
folded do ma<strong>in</strong>s con nected by flex i ble l<strong>in</strong>k ers [27]. F is<br />
kept <strong>in</strong> ac tive <strong>in</strong> a com plex with SpoIIAB. In <strong>the</strong> pres ence<br />
<strong>of</strong> unphosphorylated SpoIIAA mol e cules, SpoIIAB b<strong>in</strong>ds<br />
to SpoIIAA and ac tive F is re leased. How ever, SpoIIAB<br />
k<strong>in</strong>ase phos phory lates SpoIIAA, and <strong>the</strong> SpoIIAB-<br />
SpoIIAA com plex de cays, re leas <strong>in</strong>g so <strong>the</strong> anti- fac tor<br />
ready to block an o<strong>the</strong>r F . SpoIIAB has <strong>the</strong> ATP b<strong>in</strong>d <strong>in</strong>g<br />
Bergerat fold [28], found <strong>in</strong> histid<strong>in</strong>e k<strong>in</strong> ases and ATP-ases<br />
<strong>of</strong> <strong>the</strong> GHKL superfamily. This con sists <strong>of</strong> an / - sand -<br />
wich with a four - stranded antiparallel - sheet and three a<br />
he li ces. In <strong>the</strong> SpoIIAB mono mer 1 ex tends <strong>the</strong> four -<br />
stranded sheet. Dimerization <strong>in</strong>teractions occur among<br />
1 and 1 <strong>of</strong> each SpoIIAB mono mers. The ATP - b<strong>in</strong>d <strong>in</strong>g<br />
site is rep re sented by Asn50, which plays a key role <strong>in</strong> che -<br />
lat<strong>in</strong>g Mg 2+ , crit i cal for ATP-ase or k<strong>in</strong>ase ac tiv ity <strong>of</strong> <strong>the</strong><br />
GHKL superfamily mem bers [27]. The cat a lytic site is<br />
formed by Glu46, which func tions as a cat a lytic base <strong>in</strong> <strong>the</strong><br />
k<strong>in</strong>ase re ac tion [29]. SpoIIAA phosphorylated by SpoIIAB<br />
re quires <strong>the</strong> phosphatase ac tiv ity <strong>of</strong> SpoIIE to be come ac -<br />
tive aga<strong>in</strong>.<br />
After activation <strong>of</strong> F , which fol lows only af ter com ple -<br />
tion <strong>of</strong> <strong>the</strong> sporulation sep tum, o<strong>the</strong>r com part ment spe cific<br />
fac tors are se quen tially ac ti vated. This leads to dif fer en -<br />
ti a tion <strong>of</strong> gene ex pres sion. In <strong>the</strong> course <strong>of</strong> <strong>the</strong> sporulation<br />
pro cess, <strong>the</strong> mo<strong>the</strong>r <strong>cell</strong> en gulfs <strong>the</strong> prespore and <strong>the</strong> pro -<br />
cess <strong>of</strong> sporulation cul mi nates with pro grammed death <strong>of</strong><br />
<strong>the</strong> mo<strong>the</strong>r <strong>cell</strong>. The sur vi vor is <strong>the</strong> forespore, which ma -<br />
tures <strong>in</strong>to a very re sis tant spore able to out live hos tile en vi -<br />
ron ments. The life cy cle is com pleted by <strong>the</strong> pro cess <strong>of</strong><br />
ger mi na tion, which is <strong>in</strong>i ti ated when <strong>the</strong> free spore re turns<br />
to con di tions fa vor able for life.<br />
Future perspectives<br />
Es pe cially <strong>in</strong> ter est <strong>in</strong>g for our un der stand <strong>in</strong>g <strong>of</strong> this mech a -<br />
nism <strong>of</strong> <strong>the</strong> gene ex pres sion asym me try would be <strong>the</strong> de -<br />
tailed study <strong>of</strong> sporulation septa for ma tion by solv <strong>in</strong>g <strong>the</strong><br />
crys tal struc ture <strong>of</strong> key pro te<strong>in</strong>s <strong>of</strong> this <strong>cell</strong> di vi sion pro -<br />
cess. Al though <strong>the</strong> asym met ric <strong>cell</strong> di vi sion that oc curs<br />
dur <strong>in</strong>g sporulation dif fers from <strong>the</strong> <strong>cell</strong> di vi sion dur <strong>in</strong>g<br />
veg e ta tive growth, both pro cesses use fun da men tally <strong>the</strong><br />
same pro te<strong>in</strong> ma ch<strong>in</strong> ery [30, 31, 32]. SpoIIE pro te<strong>in</strong> is one<br />
<strong>of</strong> <strong>the</strong> most <strong>in</strong> ter est <strong>in</strong>g can di dates for crys tal lo graphic stud -<br />
ies. This large (92 kDa) pro te<strong>in</strong> con sists <strong>of</strong> three do ma<strong>in</strong>s:<br />
N-ter mi nal re gion con ta<strong>in</strong> <strong>in</strong>g 10 mem brane-span n<strong>in</strong>g he li -<br />
ces [33, 34] fol lowed by cen tral do ma<strong>in</strong>, pos si bly <strong>in</strong> volved<br />
<strong>in</strong> <strong>in</strong>termolecular <strong>in</strong> ter ac tion [22], and <strong>the</strong> C-ter mi nal do -<br />
ma<strong>in</strong>, which is spe cific pro te<strong>in</strong> phosphatase <strong>in</strong> volved <strong>in</strong> ac -<br />
ti va tion <strong>of</strong> <strong>the</strong> first forespore sigma fac tor [25]. An o<strong>the</strong>r<br />
very <strong>in</strong> ter est <strong>in</strong>g and highly stud ied sporulation spe cific<br />
pro te<strong>in</strong> from B. <strong>subtilis</strong> is DNA translocase SpoIIIE [35,<br />
36]. Re cently it has been shown that this pro te<strong>in</strong> also co op -<br />
er ates <strong>in</strong> mem brane fu sion dur <strong>in</strong>g <strong>the</strong> spore en gulf ment<br />
[37]. SpoIIIE pro te<strong>in</strong> con sists <strong>of</strong> a N-ter mi nal mem brane<br />
bound do ma<strong>in</strong> me di at <strong>in</strong>g its lo cal iza tion to <strong>the</strong> di vi sion<br />
sep tum and cytosolic C-ter mi nal do ma<strong>in</strong> ca pa ble <strong>of</strong> track -<br />
<strong>in</strong>g along DNA <strong>in</strong> <strong>the</strong> pres ence <strong>of</strong> ATP.<br />
DivIVA pro te<strong>in</strong> is an im por tant reg u la tor <strong>of</strong> <strong>cell</strong> di vi -<br />
sion dur <strong>in</strong>g veg e ta tive growth [38] and it also has been pro -<br />
posed to have a cru cial role <strong>in</strong> <strong>the</strong> sporulation pro cess by<br />
an chor <strong>in</strong>g <strong>the</strong> chro mo somes to <strong>the</strong> <strong>cell</strong> poles prior to asym -<br />
met ric di vi sion [39]. It was shown that <strong>in</strong> its na tive state<br />
DivIVA oligomerizes [40], and has a struc tural sim i lar ity<br />
to my o s<strong>in</strong> and o<strong>the</strong>r pro te<strong>in</strong>s hav <strong>in</strong>g -helical coiled-coil<br />
struc ture [41].<br />
This re view is an at tempt to sum ma rize what is known<br />
from crys tal lo graphic stud ies about <strong>the</strong> struc ture <strong>of</strong> some<br />
<strong>of</strong> <strong>the</strong> ap prox i mately 200 sporulation spe cific pro te<strong>in</strong>s <strong>of</strong> B.<br />
<strong>subtilis</strong>. The struc tures <strong>of</strong> only a small num ber <strong>of</strong> <strong>the</strong>se pro -<br />
te<strong>in</strong>s are known due to prob lems as so ci ated with <strong>the</strong> crys -<br />
tal li za tion <strong>of</strong> <strong>the</strong> pro te<strong>in</strong>s. Struc tures for many <strong>of</strong> <strong>the</strong><br />
<strong>in</strong>terest<strong>in</strong>g candidates rema<strong>in</strong> a challenge. Among <strong>the</strong>m are<br />
<strong>the</strong> mem brane bound pro te<strong>in</strong>s and pro te<strong>in</strong>s with very flex i -<br />
ble do ma<strong>in</strong>s.<br />
Acknowledgements<br />
The work <strong>in</strong> IB's lab o ra tory is sup ported by grant<br />
2/1004/21 from <strong>the</strong> Slo vak Acad emy <strong>of</strong> Sci ences and<br />
Wellcome Trust Grant 066732/Z/01/Z.<br />
Krystalografická spoleènost
I. Barák, K. Muchová, P. Florek and Z. Chromiková 17<br />
References<br />
1. M. Ji ang, W. Shao, M. Pe re go & J. A. Hoch, Mol. Micro bi -<br />
ol. 38 (2000) 535-542.<br />
2. D. Bur bu lys, K. A. Trach & J. A. Hoch, Cell 64 (1991)<br />
545-552.<br />
3. Mad hu su dan, J. Zapf, J. M.Whi te ley, J. A.Hoch, N. H. Xu -<br />
ong & K. I. Va rughe se, Structu re 4(1996) 679-690.<br />
4. A. M. Stock, J. M. Mottonen, J. B. Stock & C. E. Schutt,<br />
Na ture 337(1989) 745-749.<br />
5. C. Birck, L. Mourey, P. Gouet, B. Fabry, J. Schumacher, P.<br />
Rous seau, D. Kahn & J. P. Samama, Struc ture 7 (1999)<br />
1505-1515.<br />
6. R. J. Lewis, J. A. Brannigan, K. Muchová, I. Barák & A. J.<br />
Wilk<strong>in</strong>son, J. Mol. Biol. 294 (1999) 9-15.<br />
7. K. I. Varughese, Madhusudan, X. Z. Zhou, J. M. Whiteley<br />
& J. A. Hoch, Mol. Cell. 2 (1998) 485-493.<br />
8. J. Zapf, U. Sen, Madhusudan, J. A. Hoch & K. I.<br />
Varughese, Struc ture 8 (2000) 851-862.<br />
9. J. K. Grimsley, R. B. Tjalkens, M. A. Strauch, T. H. Bird,<br />
G. B. Spiegelman, Z. Hostomsky, J. M. Whiteley & J. A.<br />
Hoch, J. Biol. Chem. 269 (1994) 16977-16982.<br />
10. R. J. Lewis, K. Muchová, J. A. Brannigan, I. Barák, G.<br />
Leon ard & A. J. Wilk<strong>in</strong>son, J. Mol. Biol. 297 (2000)<br />
757-770.<br />
11. H. S. Cho, S. Y. Lee, D. Yan, X. Pan, J. S. Par k<strong>in</strong> son, S.<br />
Kustu, D. E. Wemmer & J. C. Pel ton, J. Mol. Biol. 297<br />
(2000) 543-551.<br />
12. R. J. Lewis, S. Kr zy w da, J. A. Bran ni gan, J. P. Tu ken burg,<br />
K. Mu cho vá, E. J. Dod son, I. Ba rák & A. J. Wil k<strong>in</strong> son,<br />
Mol. Micro bi ol. 38 (2000) 198-212.<br />
13. H. Zhao, T. Msadek, J. Zapf, Madhusudan, J. A. Hoch &<br />
K. I. Varughese, Structure 10 (2002) 1041-1050.<br />
14. R. J. Lewis, D. J. Scott, J. A. Brannigan, J. C. Ladds, M. A.<br />
Cerv<strong>in</strong>, G. B. Spiegelman, J. G. Hoggett, I. Barák & A. J.<br />
Wilk<strong>in</strong>son, J. Mol. Biol. 316 (2002) 235-245.<br />
15. I. Mandic-Mulec, L. Doukhan & I. Smith, J. Bacteriol. 177<br />
(1995) 4619-4627.<br />
16. R. J. Lewis, J. A. Brannigan, W. A. Offen, I. Smith & A. J.<br />
Wilk<strong>in</strong>son, J. Mol. Biol. 283 (1998) 907-912.<br />
17. J. Bugl<strong>in</strong>o, V. Shen, P. Hakimian & C. D. Lima, Struc ture<br />
10 (2002) 1581-1592.<br />
18. I. Barák & P. Youngman, J. Bacteriol. 178 (1996)<br />
4984-4989.<br />
19. A. Feucht, T. Magn<strong>in</strong>, M. D. Yudk<strong>in</strong> & J. Err<strong>in</strong>gton, Genes<br />
De velop. 10 (1996) 794-803.<br />
20. A. Khvorova, L. Zhang, M. L. Hig g<strong>in</strong>s & P. J. Piggot, J.<br />
Bacteriol. 180 (1998) 1256-1260.<br />
21. K. York, T. J. Kenney, S. Satola, C. P. Moran, H. Poth &<br />
P. Youngman, J. Bacteriol. 174 (1992) 2648-2658.<br />
22. I. Lucet, A. Feucht, M. D. Yudk<strong>in</strong> & J. Err<strong>in</strong>gton, EMBO J.<br />
19 (2000) 1467-1475.<br />
23. P. Prepiak, Z. Chromiková & I. Barák, Folia Microbiol.<br />
(Praha,) 46 (2001) 292-296.<br />
24. L. Duncan, S. Alper. & R. Losick, Curr. Op<strong>in</strong>. Genet. Dev.<br />
4 (1994) 630-636.<br />
25. L. Duncan, S. Alper, F. Arigoni, R. Losick & P. Stragier,<br />
Sci ence 270 (1995) 641-644.<br />
26. P. R. Seavers, R. J. Lewis, J. A. Brannigan, K. H. G.<br />
Verschueren, G. N. Murshudov & A. J. Wilk<strong>in</strong>son, Struc -<br />
ture 9 (2001) 614–615.<br />
27. E. A. Camp bell, S. Masuda, J. L. Sun, O. Muzz<strong>in</strong>, C. A.<br />
Olson, S. Wang & S. A. Darst, Cell 108 (2002) 795-807.<br />
28. A. Ber ge rat, Na tu re 386 (1997) 414-417.<br />
29. S. G. Park and M. D. Yudk<strong>in</strong>, Gene 194 (1997) 25-33.<br />
30. J. Err<strong>in</strong>gton, Curr. Op<strong>in</strong>. Microbiol. 4 (2001) 660-666.<br />
31. B. Beall and J. Lutkenhaus, Genes Dev. 5 (1991)<br />
447-455.<br />
32. K. Ireton, N. W. Gun <strong>the</strong>r & A. D. Grossman, J. Bacteriol.<br />
176 (1994) 5320-5329.<br />
33. I. Barák, J. Behari, G. Olmedo, P. Guzmán, D. P. Brown,<br />
E. Cas tro, D. Walker, J. Westphel<strong>in</strong>g & P. Youngman,<br />
Mol. Microbiol. 19 (1996) 1047-60.<br />
34. F. Arigoni, A. M. Guérout-Fleu ry, I. Barák & P. Stragier,<br />
Mol. Microbiol. 31 (1999) 1407-1715.<br />
35. L. Wu & J. Err<strong>in</strong>gton, Sci ence 264 (1994) 572-575.<br />
36. J. Bath, L. J. Wu, J. Err<strong>in</strong>g ton and J. C. Wang, Science 290<br />
(2000) 995-997.<br />
37. M. D. Sharp & K. Pogliano, Proc. Natl. Acad. Sci. USA 96<br />
(1999) 14553-14558.<br />
38. D. H. Ed wards and J. Err<strong>in</strong>gton, Mol. Microbiol. 24 (1997)<br />
905-915.<br />
39. H. B. Thomaides, M. Free man, M. E. Karoui & J.<br />
Err<strong>in</strong>gton, Genes Dev. 15 (2001) 1662-1673.<br />
40. K. Muchová, E. Kutejová, D. J. Scott, J. A. Brannigan, R.<br />
J. Lewis, A. J. Wilk<strong>in</strong>son & I. Barák,. Microbiol. 148<br />
(2002) 807-813.<br />
41. D. H. Ed wards, H. B. Thomaides & J. Err<strong>in</strong>gton, EMBO J.<br />
19 (2000) 2719-2727.<br />
Krystalografická spoleènost
18 ABSTRACTS<br />
Krystalografická spoleènost