Optimizing Sorting with Genetic Algorithms - Polaris
Optimizing Sorting with Genetic Algorithms - Polaris
Optimizing Sorting with Genetic Algorithms - Polaris
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
AMD Sun SGI Intel Intel<br />
CPU Athlon MP UltraSparcIIIi R12000 Itanium 2 Pentium 4<br />
Frequency 1.2GHz 1GHz 300MHz 1.5GHz 2GHz<br />
L1d/L1i Cache 128KB 64KB/32KB 32KB/32KB 16KB/16KB 8KB/12KB<br />
L2 Cache 256KB 1MB 4MB 256KB 512KB<br />
Memory 1GB 4GB 1GB 8GB 512KB<br />
OS RedHat9 SunOS5.8 IRIX64 v6.5 RedHat7.2 RedHat7.2<br />
Compiler gcc3.2.2 Workshop cc 5.0 MIPSPro cc 7.3.0 gcc3.3.2 gcc3.3.1<br />
Options -O3 -native -xO5 -O3 -TARG: -O3 -O3<br />
platform=IP30<br />
Table 2: Test Platforms. Intel Pentium 4 has a 8KB trace cache instead of a L1 instruction cache. Intel Itanium2 has a L3 cache of<br />
6MB.<br />
Execution Time (Cycles per key)<br />
Execution Time (Cycles per key)<br />
800<br />
700<br />
600<br />
500<br />
400<br />
300<br />
200<br />
Intel Pentium 4<br />
100<br />
100 1000 10000 100000 1e+06 1e+07<br />
1000<br />
900<br />
800<br />
700<br />
600<br />
500<br />
400<br />
300<br />
200<br />
Quicksort<br />
CC-radix<br />
Standard Deviation<br />
Multi-way Merge<br />
Gene Sort<br />
Intel MKL<br />
C++ STL<br />
100<br />
100 1000 10000 100000 1e+06 1e+07<br />
Quicksort<br />
CC-radix<br />
Intel Itanium 2<br />
Standard Deviation<br />
Multi-way Merge<br />
Gene Sort<br />
Intel MKL<br />
C++ STL<br />
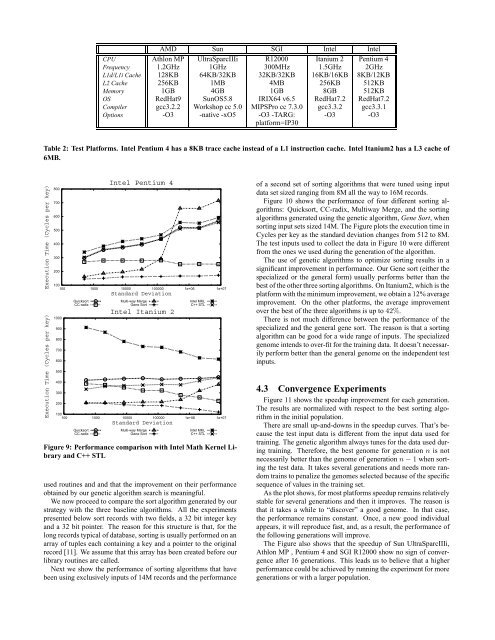
Figure 9: Performance comparison <strong>with</strong> Intel Math Kernel Library<br />
and C++ STL<br />
used routines and and that the improvement on their performance<br />
obtained by our genetic algorithm search is meaningful.<br />
We now proceed to compare the sort algorithm generated by our<br />
strategy <strong>with</strong> the three baseline algorithms. All the experiments<br />
presented below sort records <strong>with</strong> two fields, a 32 bit integer key<br />
and a 32 bit pointer. The reason for this structure is that, for the<br />
long records typical of database, sorting is usually performed on an<br />
array of tuples each containing a key and a pointer to the original<br />
record [11]. We assume that this array has been created before our<br />
library routines are called.<br />
Next we show the performance of sorting algorithms that have<br />
been using exclusively inputs of 14M records and the performance<br />
of a second set of sorting algorithms that were tuned using input<br />
data set sized ranging from 8M all the way to 16M records.<br />
Figure 10 shows the performance of four different sorting algorithms:<br />
Quicksort, CC-radix, Multiway Merge, and the sorting<br />
algorithms generated using the genetic algorithm, Gene Sort, when<br />
sorting input sets sized 14M. The Figure plots the execution time in<br />
Cycles per key as the standard deviation changes from 512 to 8M.<br />
The test inputs used to collect the data in Figure 10 were different<br />
from the ones we used during the generation of the algorithm.<br />
The use of genetic algorithms to optimize sorting results in a<br />
significant improvement in performance. Our Gene sort (either the<br />
specialized or the general form) usually performs better than the<br />
best of the other three sorting algorithms. On Itanium2, which is the<br />
platform <strong>with</strong> the minimum improvement, we obtain a 12% average<br />
improvement. On the other platforms, the average improvement<br />
over the best of the three algorithms is up to 42%.<br />
There is not much difference between the performance of the<br />
specialized and the general gene sort. The reason is that a sorting<br />
algorithm can be good for a wide range of inputs. The specialized<br />
genome intends to over-fit for the training data. It doesn’t necessarily<br />
perform better than the general genome on the independent test<br />
inputs.<br />
4.3 Convergence Experiments<br />
Figure 11 shows the speedup improvement for each generation.<br />
The results are normalized <strong>with</strong> respect to the best sorting algorithm<br />
in the initial population.<br />
There are small up-and-downs in the speedup curves. That’s because<br />
the test input data is different from the input data used for<br />
training. The genetic algorithm always tunes for the data used during<br />
training. Therefore, the best genome for generation n is not<br />
necessarily better than the genome of generation n − 1 when sorting<br />
the test data. It takes several generations and needs more random<br />
trains to penalize the genomes selected because of the specific<br />
sequence of values in the training set.<br />
As the plot shows, for most platforms speedup remains relatively<br />
stable for several generations and then it improves. The reason is<br />
that it takes a while to “discover” a good genome. In that case,<br />
the performance remains constant. Once, a new good individual<br />
appears, it will reproduce fast, and, as a result, the performance of<br />
the following generations will improve.<br />
The Figure also shows that the speedup of Sun UltraSparcIIIi,<br />
Athlon MP , Pentium 4 and SGI R12000 show no sign of convergence<br />
after 16 generations. This leads us to believe that a higher<br />
performance could be achieved by running the experiment for more<br />
generations or <strong>with</strong> a larger population.