The impact of Ty3-gypsy group LTR retrotransposons ... - URGV - Inra
The impact of Ty3-gypsy group LTR retrotransposons ... - URGV - Inra
The impact of Ty3-gypsy group LTR retrotransposons ... - URGV - Inra
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
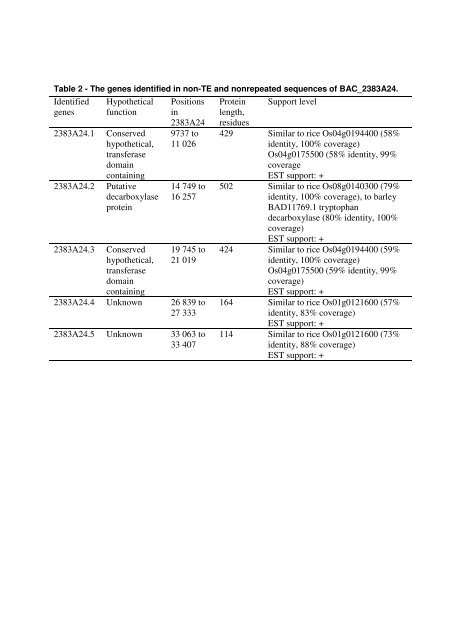
Table 2 - <strong>The</strong> genes identified in non-TE and nonrepeated sequences <strong>of</strong> BAC_2383A24.<br />
Identified<br />
genes<br />
Hypothetical<br />
function<br />
Positions<br />
in<br />
Protein<br />
length,<br />
Support level<br />
2383A24.1 Conserved<br />
hypothetical,<br />
transferase<br />
domain<br />
containing<br />
2383A24.2 Putative<br />
decarboxylase<br />
protein<br />
2383A24.3 Conserved<br />
hypothetical,<br />
transferase<br />
domain<br />
containing<br />
2383A24<br />
9737 to<br />
11 026<br />
14 749 to<br />
16 257<br />
19 745 to<br />
21 019<br />
2383A24.4 Unknown 26 839 to<br />
27 333<br />
2383A24.5 Unknown 33 063 to<br />
33 407<br />
residues<br />
429 Similar to rice Os04g0194400 (58%<br />
identity, 100% coverage)<br />
Os04g0175500 (58% identity, 99%<br />
coverage<br />
EST support: +<br />
502 Similar to rice Os08g0140300 (79%<br />
identity, 100% coverage), to barley<br />
BAD11769.1 tryptophan<br />
decarboxylase (80% identity, 100%<br />
coverage)<br />
EST support: +<br />
424 Similar to rice Os04g0194400 (59%<br />
identity, 100% coverage)<br />
Os04g0175500 (59% identity, 99%<br />
coverage)<br />
EST support: +<br />
164 Similar to rice Os01g0121600 (57%<br />
identity, 83% coverage)<br />
EST support: +<br />
114 Similar to rice Os01g0121600 (73%<br />
identity, 88% coverage)<br />
EST support: +