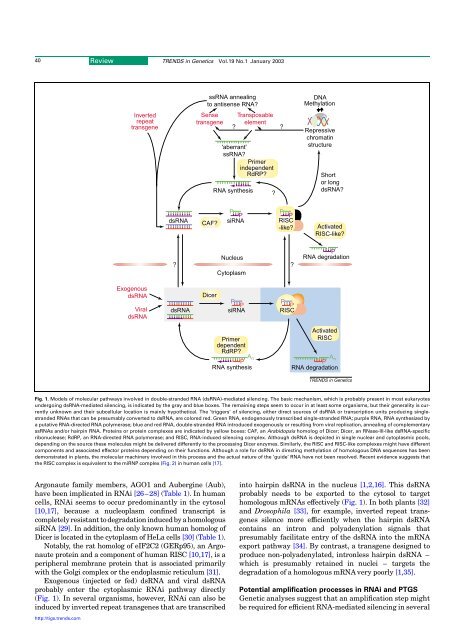
40Review TRENDS <strong>in</strong> Genetics Vol.19 No.1 January 2003ss<strong>RNA</strong> anneal<strong>in</strong>gto antisense <strong>RNA</strong>?DNAMethylationInvertedrepeattransgeneSensetransgene?Transposableelement'aberrant'ss<strong>RNA</strong>?Primer<strong>in</strong>dependentRdRP?<strong>RNA</strong> syn<strong>the</strong>sis??Repressivechromat<strong>in</strong>structureShortor longds<strong>RNA</strong>?PPPPds<strong>RNA</strong> CAF? si<strong>RNA</strong> RISC-like?ActivatedRISC-like??NucleusCytoplasm?P<strong>RNA</strong> degradationExogenousds<strong>RNA</strong>Viralds<strong>RNA</strong>DicerPPPPds<strong>RNA</strong> si<strong>RNA</strong> RISCActivatedPrimerRISCdependentRdRP?An A nPP<strong>RNA</strong> syn<strong>the</strong>sis<strong>RNA</strong> degradationTRENDS <strong>in</strong> GeneticsFig. 1. Models of molecular pathways <strong>in</strong>volved <strong>in</strong> double-str<strong>and</strong>ed <strong>RNA</strong> (ds<strong>RNA</strong>)-mediated silenc<strong>in</strong>g. The basic mechanism, which is probably present <strong>in</strong> most eukaryotesundergo<strong>in</strong>g ds<strong>RNA</strong>-mediated silenc<strong>in</strong>g, is <strong>in</strong>dicated by <strong>the</strong> gray <strong>and</strong> blue boxes. The rema<strong>in</strong><strong>in</strong>g steps seem to occur <strong>in</strong> at least some organisms, but <strong>the</strong>ir generality is currentlyunknown <strong>and</strong> <strong>the</strong>ir sub<strong>cell</strong>ular location is ma<strong>in</strong>ly hypo<strong>the</strong>tical. The ‘triggers’ of silenc<strong>in</strong>g, ei<strong>the</strong>r direct sources of ds<strong>RNA</strong> or transcription units produc<strong>in</strong>g s<strong>in</strong>glestr<strong>and</strong>ed<strong>RNA</strong>s that can be presumably converted to ds<strong>RNA</strong>, are colored red. Green <strong>RNA</strong>, endogenously transcribed s<strong>in</strong>gle-str<strong>and</strong>ed <strong>RNA</strong>; purple <strong>RNA</strong>, <strong>RNA</strong> syn<strong>the</strong>sized bya putative <strong>RNA</strong>-directed <strong>RNA</strong> polymerase; blue <strong>and</strong> red <strong>RNA</strong>, double-str<strong>and</strong>ed <strong>RNA</strong> <strong>in</strong>troduced exogenously or result<strong>in</strong>g from viral replication, anneal<strong>in</strong>g of complementaryss<strong>RNA</strong>s <strong>and</strong>/or hairp<strong>in</strong> <strong>RNA</strong>. Prote<strong>in</strong>s or prote<strong>in</strong> complexes are <strong>in</strong>dicated by yellow boxes: CAF, an Arabidopsis homolog of Dicer; Dicer, an RNase-III-like ds<strong>RNA</strong>-specificribonuclease; RdRP, an <strong>RNA</strong>-directed <strong>RNA</strong> polymerase; <strong>and</strong> RISC, <strong>RNA</strong>-<strong>in</strong>duced silenc<strong>in</strong>g complex. Although ds<strong>RNA</strong> is depicted <strong>in</strong> s<strong>in</strong>gle nuclear <strong>and</strong> cytoplasmic pools,depend<strong>in</strong>g on <strong>the</strong> source <strong>the</strong>se molecules might be delivered differently to <strong>the</strong> process<strong>in</strong>g Dicer enzymes. Similarly, <strong>the</strong> RISC <strong>and</strong> RISC-like complexes might have differentcomponents <strong>and</strong> associated effector prote<strong>in</strong>s depend<strong>in</strong>g on <strong>the</strong>ir <strong>functions</strong>. Although a role for ds<strong>RNA</strong> <strong>in</strong> direct<strong>in</strong>g methylation of homologous DNA sequences has beendemonstrated <strong>in</strong> plants, <strong>the</strong> molecular mach<strong>in</strong>ery <strong>in</strong>volved <strong>in</strong> this process <strong>and</strong> <strong>the</strong> actual nature of <strong>the</strong> ‘guide’ <strong>RNA</strong> have not been resolved. Recent evidence suggests that<strong>the</strong> RISC complex is equivalent to <strong>the</strong> miRNP complex (Fig. 2) <strong>in</strong> human <strong>cell</strong>s [17].Argonaute family members, AGO1 <strong>and</strong> Auberg<strong>in</strong>e (Aub),have been implicated <strong>in</strong> <strong>RNA</strong>i [26–28] (Table 1). In human<strong>cell</strong>s, <strong>RNA</strong>i seems to occur predom<strong>in</strong>antly <strong>in</strong> <strong>the</strong> cytosol[10,17], because a nucleoplasm conf<strong>in</strong>ed transcript iscompletely resistant to degradation <strong>in</strong>duced by a homologoussi<strong>RNA</strong> [29]. In addition, <strong>the</strong> only known human homolog ofDicer is located <strong>in</strong> <strong>the</strong> cytoplasm of HeLa <strong>cell</strong>s [30] (Table 1).Notably, <strong>the</strong> rat homolog of eIF2C2 (GERp95), an Argonauteprote<strong>in</strong> <strong>and</strong> a component of human RISC [10,17],isaperipheral membrane prote<strong>in</strong> that is associated primarilywith <strong>the</strong> Golgi complex or <strong>the</strong> endoplasmic reticulum [31].Exogenous (<strong>in</strong>jected or fed) ds<strong>RNA</strong> <strong>and</strong> viral ds<strong>RNA</strong>probably enter <strong>the</strong> cytoplasmic <strong>RNA</strong>i pathway directly(Fig. 1). In several organisms, however, <strong>RNA</strong>i can also be<strong>in</strong>duced by <strong>in</strong>verted repeat transgenes that are transcribed<strong>in</strong>to hairp<strong>in</strong> ds<strong>RNA</strong> <strong>in</strong> <strong>the</strong> nucleus [1,2,16]. Thisds<strong>RNA</strong>probably needs to be exported to <strong>the</strong> cytosol to targethomologous m<strong>RNA</strong>s effectively (Fig. 1). In both plants [32]<strong>and</strong> Drosophila [33], for example, <strong>in</strong>verted repeat transgenessilence more efficiently when <strong>the</strong> hairp<strong>in</strong> ds<strong>RNA</strong>conta<strong>in</strong>s an <strong>in</strong>tron <strong>and</strong> polyadenylation signals thatpresumably facilitate entry of <strong>the</strong> ds<strong>RNA</strong> <strong>in</strong>to <strong>the</strong> m<strong>RNA</strong>export pathway [34]. By contrast, a transgene designed toproduce non-polyadenylated, <strong>in</strong>tronless hairp<strong>in</strong> ds<strong>RNA</strong> –which is presumably reta<strong>in</strong>ed <strong>in</strong> nuclei – targets <strong>the</strong>degradation of a homologous m<strong>RNA</strong> very poorly [1,35].Potential amplification processes <strong>in</strong> <strong>RNA</strong>i <strong>and</strong> PTGSGenetic analyses suggest that an amplification step mightbe required for efficient <strong>RNA</strong>-mediated silenc<strong>in</strong>g <strong>in</strong> severalhttp://tigs.trends.com
Review TRENDS <strong>in</strong> Genetics Vol.19 No.1 January 2003 41Table 1. Genetic <strong>and</strong>/or biochemical components of <strong>the</strong> <strong>RNA</strong>i/PTGS mach<strong>in</strong>ery discussed <strong>in</strong> <strong>the</strong> text a,bGene function A. thaliana C. elegans C. re<strong>in</strong>hardtii D. discoideum D. melanogaster H. sapiens N. crassa S. pombeRNase III þ <strong>RNA</strong> helicase CAF DCR-1 Dicer Dicer [C] DCR1Unknown (Argonaute) AGO1 RDE-1,AGO1 [C], eIF2C1, QDE2 AGO1ALG-1,ALG-2AGO2,AUB [C],PIWI [N]eIF2C2Putative RdRP SDE1/SGS2 RRF-1,RRPA QDE1 RDP1 [N]RRF-3<strong>RNA</strong> helicaseDRH-1,DRH2MUT6 [N]Gem<strong>in</strong>3[N þ C]ds<strong>RNA</strong>-b<strong>in</strong>d<strong>in</strong>g prote<strong>in</strong>RDE-4RNase D likeMUT-7RecQ DNA helicaseQDE3UnknownGem<strong>in</strong>4[N þ C]a This list is not comprehensive <strong>and</strong> only some prote<strong>in</strong>s are <strong>in</strong>dicated for each of <strong>the</strong> follow<strong>in</strong>g organisms: Arabidopsis thaliana, Caenorhabditis elegans, Chlamydomonasre<strong>in</strong>hardtii, Dictyostelium discoideum, Drosophila melanogaster, Homo sapiens, Neurospora crassa, Schizosaccharomyces pombe.b Where known, <strong>the</strong> sub<strong>cell</strong>ular localization of <strong>the</strong> prote<strong>in</strong>s is shown <strong>in</strong> paren<strong>the</strong>ses: C, cytoplasmic; N, nuclear.systems. Recent studies <strong>in</strong> C. elegans have led to a model <strong>in</strong>which primary si<strong>RNA</strong>s (derived from <strong>the</strong> trigger ds<strong>RNA</strong>)might prime <strong>the</strong> syn<strong>the</strong>sis of additional ds<strong>RNA</strong>, us<strong>in</strong>g <strong>the</strong>target m<strong>RNA</strong> as a template, <strong>in</strong> a reaction catalyzed by aputative <strong>RNA</strong>-directed <strong>RNA</strong> polymerase (RdRP) [4,36](Fig. 1, cytoplasm). The newly syn<strong>the</strong>sized ds<strong>RNA</strong> would<strong>the</strong>n be cleaved by Dicer to generate secondary si<strong>RNA</strong>s at asufficient concentration to achieve efficient target m<strong>RNA</strong>degradation by RISC [4,36]. In support of this model, <strong>the</strong><strong>in</strong>jection of short antisense <strong>RNA</strong> oligomers <strong>in</strong>to C. eleganscan trigger silenc<strong>in</strong>g of endogenous genes, <strong>and</strong> this effect isdependent on a functional Dicer (DCR-1) [37].Putative homologs of a tomato RdRP are required forPTGS triggered by sense transgenes <strong>in</strong> Arabidopsis, forquell<strong>in</strong>g <strong>in</strong> Neurospora, <strong>and</strong> for <strong>RNA</strong>i <strong>in</strong> C. elegans <strong>and</strong>Dictyostelium discoideum [1–4,36,38–40]; however, biochemicalactivity rema<strong>in</strong>s to be demonstrated def<strong>in</strong>itivelyfor any of <strong>the</strong> RdRP homologs implicated <strong>in</strong> <strong>RNA</strong>i <strong>and</strong>PTGS [5]. In addition, <strong>in</strong> both Drosophila <strong>and</strong> humans, <strong>the</strong>results of many experiments argue aga<strong>in</strong>st an obligatoryrole for an RdRP <strong>in</strong> ds<strong>RNA</strong>-<strong>in</strong>duced <strong>RNA</strong>i [5,9,12],although an RdRP-like activity has been reported <strong>in</strong>extracts from Drosophila embryos [41].Because si<strong>RNA</strong> production <strong>and</strong> target m<strong>RNA</strong> cleavagemost probably occur <strong>in</strong> <strong>the</strong> cytosol, it is tempt<strong>in</strong>g tospeculate that <strong>the</strong> RdRP-dependent amplification stepdescribed <strong>in</strong> C. elegans also occurs <strong>in</strong> <strong>the</strong> cytoplasm (Fig. 1).Notably, RDE-1 <strong>and</strong> RDE-4 are required to <strong>in</strong>itiate <strong>RNA</strong>i<strong>in</strong>duced by <strong>in</strong>jected or transgene-produced long ds<strong>RNA</strong>s[20], but not for silenc<strong>in</strong>g triggered by short antisense<strong>RNA</strong>s [37]. Thus, <strong>the</strong> RdRP-generated ds<strong>RNA</strong> might bedelivered to Dicer differently to <strong>the</strong> way <strong>in</strong> whichexogenous or transgenic ds<strong>RNA</strong> is delivered [19] (Fig. 1,cytoplasm). In Dictyostelium, RRPA, a putative RdRPrequired for <strong>RNA</strong>i, conta<strong>in</strong>s an N-term<strong>in</strong>al <strong>RNA</strong> helicasedoma<strong>in</strong> that has not been found <strong>in</strong> o<strong>the</strong>r RdRPs [40].Intrigu<strong>in</strong>gly, <strong>the</strong> closest homolog of this helicase doma<strong>in</strong> isthat of C. elegans Dicer, suggest<strong>in</strong>g that doma<strong>in</strong> swapp<strong>in</strong>gmight have occurred between Dictyostelium Dicer <strong>and</strong>RRPA [40]. If this is true, <strong>the</strong>n perhaps Dicer <strong>and</strong> an RdRP<strong>in</strong>teract <strong>in</strong> a complex such that <strong>the</strong> ds<strong>RNA</strong> generated byRdRP activity is immediately accessible <strong>and</strong> rapidlyprocessed by Dicer.In C. elegans, RDE-1, RDE-4 <strong>and</strong> RRF-1 (a putativeRdRP) are required for ds<strong>RNA</strong>-<strong>in</strong>duced silenc<strong>in</strong>g <strong>in</strong> <strong>the</strong>soma, even when <strong>the</strong> trigger ds<strong>RNA</strong> is expressed from atransgene <strong>in</strong> <strong>the</strong> nuclei of target tissues [16,19,36]. SDE1/SGS2 (a putative RdRP) <strong>and</strong> AGO1 (<strong>the</strong> first describedmember of <strong>the</strong> Argonaute family [15]) are also requiredfor PTGS <strong>in</strong>duced by sense transgenes <strong>in</strong> Arabidopsis[38,39,42]. They are, however, completely dispensable forPTGS <strong>in</strong>duced by <strong>the</strong> expression of <strong>in</strong>verted repeat transgenesthat produce hairp<strong>in</strong> ds<strong>RNA</strong> [42]. This suggests that<strong>the</strong> putative amplification step catalyzed by ArabidopsisSDE1/SGS2 might be different to that mediated byC. elegans RRF-1 [42].In plants, <strong>the</strong>re is also an SDE1/SGS2-dependentspread<strong>in</strong>g of silenc<strong>in</strong>g from <strong>the</strong> region homologous to <strong>the</strong>trigger ds<strong>RNA</strong> <strong>in</strong>to <strong>the</strong> adjacent non-homologous 5 0 <strong>and</strong> 3 0regions of a target transgene [43]. This is not consistentwith <strong>the</strong> simple notion that primary si<strong>RNA</strong>s prime 5 0 ! 3 0ds<strong>RNA</strong> syn<strong>the</strong>sis on sense m<strong>RNA</strong>. A possible explanationis that trigger ds<strong>RNA</strong>s or si<strong>RNA</strong>s <strong>in</strong>teract with <strong>the</strong>transgene DNA <strong>and</strong> <strong>in</strong>duce changes <strong>in</strong> chromat<strong>in</strong> structure[38,43] (Fig. 1, nucleus, repressive chromat<strong>in</strong> structure).This would lead to <strong>the</strong> production of ‘aberrant’, butnearly full-length transgenic <strong>RNA</strong>s that could be used bySDE1/SGS2 for primer-<strong>in</strong>dependent amplification [38,43](Fig. 1, nucleus, ‘aberrant ss<strong>RNA</strong>?’).In several plant species, PTGS of transgenes usuallycorrelates with DNA methylation <strong>in</strong> transcribed regions[1–3,44]. Chromat<strong>in</strong> modifications associated with (orpreced<strong>in</strong>g) DNA methylation might lead to <strong>the</strong> productionof truncated aberrant <strong>RNA</strong>s, as has been reported <strong>in</strong>filamentous fungi [45]. A role for DNA methylation <strong>and</strong>/orchromat<strong>in</strong> modification <strong>in</strong> PTGS of sense transgenes isalso supported by <strong>the</strong> observation that mutations <strong>in</strong>MET1/DDM2 (a DNA methyltransferase) <strong>and</strong> <strong>in</strong> DDM1(a member of <strong>the</strong> SWI2/SNF2 family of chromat<strong>in</strong>remodel<strong>in</strong>gprote<strong>in</strong>s) can affect both <strong>the</strong> degree <strong>and</strong>persistence of post-transcriptional silenc<strong>in</strong>g <strong>in</strong> Arabidopsis[44], although <strong>in</strong>direct effects, such as saturation of <strong>the</strong>PTGS mach<strong>in</strong>ery by <strong>the</strong> unregulated expression ofendogenous <strong>RNA</strong>s, cannot be ruled out.The nature of <strong>the</strong> postulated aberrant <strong>RNA</strong> rema<strong>in</strong>sunknown [1,2,5,38], but PTGS can be <strong>in</strong>duced <strong>in</strong> soybeanhttp://tigs.trends.com