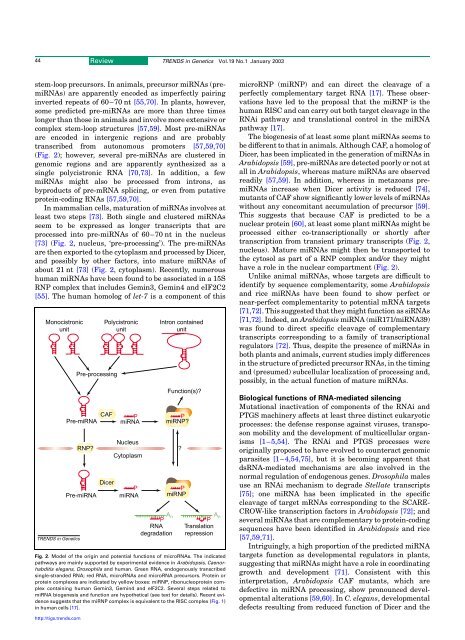
44Review TRENDS <strong>in</strong> Genetics Vol.19 No.1 January 2003stem-loop precursors. In animals, precursor mi<strong>RNA</strong>s (premi<strong>RNA</strong>s)are apparently encoded as imperfectly pair<strong>in</strong>g<strong>in</strong>verted repeats of 60–70 nt [55,70]. In plants, however,some predicted pre-mi<strong>RNA</strong>s are more than three timeslonger than those <strong>in</strong> animals <strong>and</strong> <strong>in</strong>volve more extensive orcomplex stem-loop structures [57,59]. Most pre-mi<strong>RNA</strong>sare encoded <strong>in</strong> <strong>in</strong>tergenic regions <strong>and</strong> are probablytranscribed from autonomous promoters [57,59,70](Fig. 2); however, several pre-mi<strong>RNA</strong>s are clustered <strong>in</strong>genomic regions <strong>and</strong> are apparently syn<strong>the</strong>sized as as<strong>in</strong>gle polycistronic <strong>RNA</strong> [70,73]. In addition, a fewmi<strong>RNA</strong>s might also be processed from <strong>in</strong>trons, asbyproducts of pre-m<strong>RNA</strong> splic<strong>in</strong>g, or even from putativeprote<strong>in</strong>-cod<strong>in</strong>g <strong>RNA</strong>s [57,59,70].In mammalian <strong>cell</strong>s, maturation of mi<strong>RNA</strong>s <strong>in</strong>volves atleast two steps [73]. Both s<strong>in</strong>gle <strong>and</strong> clustered mi<strong>RNA</strong>sseem to be expressed as longer transcripts that areprocessed <strong>in</strong>to pre-mi<strong>RNA</strong>s of 60–70 nt <strong>in</strong> <strong>the</strong> nucleus[73] (Fig. 2, nucleus, ‘pre-process<strong>in</strong>g’). The pre-mi<strong>RNA</strong>sare <strong>the</strong>n exported to <strong>the</strong> cytoplasm <strong>and</strong> processed by Dicer,<strong>and</strong> possibly by o<strong>the</strong>r factors, <strong>in</strong>to mature mi<strong>RNA</strong>s ofabout 21 nt [73] (Fig. 2, cytoplasm). Recently, numeroushuman mi<strong>RNA</strong>s have been found to be associated <strong>in</strong> a 15SRNP complex that <strong>in</strong>cludes Gem<strong>in</strong>3, Gem<strong>in</strong>4 <strong>and</strong> eIF2C2[55]. The human homolog of let-7 is a component of thisMonocistronicunitTRENDS <strong>in</strong> GeneticsPre-process<strong>in</strong>gCAFPre-mi<strong>RNA</strong>RNP?Pre-mi<strong>RNA</strong>PolycistronicunitDicerPmi<strong>RNA</strong>NucleusCytoplasmPmi<strong>RNA</strong>A nP<strong>RNA</strong>degradationIntron conta<strong>in</strong>edunitFunction(s)?PmiRNP?miRNP PA nPTranslationrepressionFig. 2. Model of <strong>the</strong> orig<strong>in</strong> <strong>and</strong> potential <strong>functions</strong> of micro<strong>RNA</strong>s. The <strong>in</strong>dicatedpathways are ma<strong>in</strong>ly supported by experimental evidence <strong>in</strong> Arabidopsis, Caenorhabditiselegans, Drosophila <strong>and</strong> human. Green <strong>RNA</strong>, endogenously transcribeds<strong>in</strong>gle-str<strong>and</strong>ed <strong>RNA</strong>; red <strong>RNA</strong>, micro<strong>RNA</strong>s <strong>and</strong> micro<strong>RNA</strong> precursors. Prote<strong>in</strong> orprote<strong>in</strong> complexes are <strong>in</strong>dicated by yellow boxes: miRNP, ribonucleoprote<strong>in</strong> complexconta<strong>in</strong><strong>in</strong>g human Gem<strong>in</strong>3, Gem<strong>in</strong>4 <strong>and</strong> eIF2C2. Several steps related tomi<strong>RNA</strong> biogenesis <strong>and</strong> function are hypo<strong>the</strong>tical (see text for details). Recent evidencesuggests that <strong>the</strong> miRNP complex is equivalent to <strong>the</strong> RISC complex (Fig. 1)<strong>in</strong> human <strong>cell</strong>s [17].?microRNP (miRNP) <strong>and</strong> can direct <strong>the</strong> cleavage of aperfectly complementary target <strong>RNA</strong> [17]. These observationshave led to <strong>the</strong> proposal that <strong>the</strong> miRNP is <strong>the</strong>human RISC <strong>and</strong> can carry out both target cleavage <strong>in</strong> <strong>the</strong><strong>RNA</strong>i pathway <strong>and</strong> translational control <strong>in</strong> <strong>the</strong> mi<strong>RNA</strong>pathway [17].The biogenesis of at least some plant mi<strong>RNA</strong>s seems tobe different to that <strong>in</strong> animals. Although CAF, a homolog ofDicer, has been implicated <strong>in</strong> <strong>the</strong> generation of mi<strong>RNA</strong>s <strong>in</strong>Arabidopsis [59], pre-mi<strong>RNA</strong>s are detected poorly or not atall <strong>in</strong> Arabidopsis, whereas mature mi<strong>RNA</strong>s are observedreadily [57,59]. In addition, whereas <strong>in</strong> metazoans premi<strong>RNA</strong>s<strong>in</strong>crease when Dicer activity is reduced [74],mutants of CAF show significantly lower levels of mi<strong>RNA</strong>swithout any concomitant accumulation of precursor [59].This suggests that because CAF is predicted to be anuclear prote<strong>in</strong> [60], at least some plant mi<strong>RNA</strong>s might beprocessed ei<strong>the</strong>r co-transcriptionally or shortly aftertranscription from transient primary transcripts (Fig. 2,nucleus). Mature mi<strong>RNA</strong>s might <strong>the</strong>n be transported to<strong>the</strong> cytosol as part of a RNP complex <strong>and</strong>/or <strong>the</strong>y mighthave a role <strong>in</strong> <strong>the</strong> nuclear compartment (Fig. 2).Unlike animal mi<strong>RNA</strong>s, whose targets are difficult toidentify by sequence complementarity, some Arabidopsis<strong>and</strong> rice mi<strong>RNA</strong>s have been found to show perfect ornear-perfect complementarity to potential m<strong>RNA</strong> targets[71,72]. This suggested that <strong>the</strong>y might function as si<strong>RNA</strong>s[71,72]. Indeed, an Arabidopsis mi<strong>RNA</strong> (miR171/mi<strong>RNA</strong>39)was found to direct specific cleavage of complementarytranscripts correspond<strong>in</strong>g to a family of transcriptionalregulators [72]. Thus, despite <strong>the</strong> presence of mi<strong>RNA</strong>s <strong>in</strong>both plants <strong>and</strong> animals, current studies imply differences<strong>in</strong> <strong>the</strong> structure of predicted precursor <strong>RNA</strong>s, <strong>in</strong> <strong>the</strong> tim<strong>in</strong>g<strong>and</strong> (presumed) sub<strong>cell</strong>ular localization of process<strong>in</strong>g <strong>and</strong>,possibly, <strong>in</strong> <strong>the</strong> actual function of mature mi<strong>RNA</strong>s.Biological <strong>functions</strong> of <strong>RNA</strong>-mediated silenc<strong>in</strong>gMutational <strong>in</strong>activation of components of <strong>the</strong> <strong>RNA</strong>i <strong>and</strong>PTGS mach<strong>in</strong>ery affects at least three dist<strong>in</strong>ct eukaryoticprocesses: <strong>the</strong> defense response aga<strong>in</strong>st viruses, transposonmobility <strong>and</strong> <strong>the</strong> development of multi<strong>cell</strong>ular organisms[1–5,54]. The <strong>RNA</strong>i <strong>and</strong> PTGS processes wereorig<strong>in</strong>ally proposed to have evolved to counteract genomicparasites [1–4,54,75], but it is becom<strong>in</strong>g apparent thatds<strong>RNA</strong>-mediated mechanisms are also <strong>in</strong>volved <strong>in</strong> <strong>the</strong>normal regulation of endogenous genes. Drosophila malesuse an <strong>RNA</strong>i mechanism to degrade Stellate transcripts[75]; one mi<strong>RNA</strong> has been implicated <strong>in</strong> <strong>the</strong> specificcleavage of target m<strong>RNA</strong>s correspond<strong>in</strong>g to <strong>the</strong> SCARE-CROW-like transcription factors <strong>in</strong> Arabidopsis [72]; <strong>and</strong>several mi<strong>RNA</strong>s that are complementary to prote<strong>in</strong>-cod<strong>in</strong>gsequences have been identified <strong>in</strong> Arabidopsis <strong>and</strong> rice[57,59,71].Intrigu<strong>in</strong>gly, a high proportion of <strong>the</strong> predicted mi<strong>RNA</strong>targets function as developmental regulators <strong>in</strong> plants,suggest<strong>in</strong>g that mi<strong>RNA</strong>s might have a role <strong>in</strong> coord<strong>in</strong>at<strong>in</strong>ggrowth <strong>and</strong> development [71]. Consistent with this<strong>in</strong>terpretation, Arabidopsis CAF mutants, which aredefective <strong>in</strong> mi<strong>RNA</strong> process<strong>in</strong>g, show pronounced developmentalalterations [59,60].InC. elegans, developmentaldefects result<strong>in</strong>g from reduced function of Dicer <strong>and</strong> <strong>the</strong>http://tigs.trends.com
Review TRENDS <strong>in</strong> Genetics Vol.19 No.1 January 2003 45Argonaute-like prote<strong>in</strong>s ALG-1 <strong>and</strong> ALG-2 have also beenattributed to <strong>the</strong> improper process<strong>in</strong>g of mi<strong>RNA</strong> precursors<strong>and</strong> a reduction <strong>in</strong> mature st<strong>RNA</strong> expression [3,74].Itis <strong>the</strong>refore tempt<strong>in</strong>g to propose <strong>the</strong> existence of ancient,mi<strong>RNA</strong>-mediated mechanisms that regulate endogenousgenes <strong>in</strong> eukaryotes. But most endogenous small <strong>RNA</strong>scloned from animals, <strong>and</strong> several from plants, do notmatch prote<strong>in</strong>-cod<strong>in</strong>g or structural <strong>RNA</strong>s <strong>and</strong> <strong>the</strong>irmechanistic roles, with <strong>the</strong> exception of C. elegans st<strong>RNA</strong>s,rema<strong>in</strong> unknown [55,70,74].Dicer processes precursor ds<strong>RNA</strong>s to make both si<strong>RNA</strong>s<strong>and</strong> mi<strong>RNA</strong>s. In organisms encod<strong>in</strong>g only one Dicer, as<strong>in</strong>gle pathway might h<strong>and</strong>le small <strong>RNA</strong>s. In o<strong>the</strong>r words,<strong>the</strong> mi<strong>RNA</strong> <strong>and</strong> si<strong>RNA</strong> pathways might be <strong>in</strong>terchangeablefrom biogenesis of <strong>the</strong> small <strong>RNA</strong> to <strong>in</strong>teraction withits target. The f<strong>in</strong>al outcome, such as m<strong>RNA</strong> degradationor, for example, translational repression, would depend on<strong>the</strong> degree of complementarity to <strong>the</strong> target <strong>RNA</strong> <strong>and</strong>,presumably, on associated effector prote<strong>in</strong>s [3,17]. Thismodel is consistent with <strong>the</strong> f<strong>in</strong>d<strong>in</strong>gs that short hairp<strong>in</strong><strong>RNA</strong>s, resembl<strong>in</strong>g mi<strong>RNA</strong> precursors, can <strong>in</strong>duce <strong>RNA</strong>i onperfectly homologous target m<strong>RNA</strong>s [3,17], <strong>and</strong> that <strong>the</strong>human RISC seems to be equivalent to <strong>the</strong> 15S miRNPthat is associated with many mi<strong>RNA</strong>s [17,55]. Alternatively,two dist<strong>in</strong>ct pathways, <strong>in</strong>tersect<strong>in</strong>g at <strong>the</strong> Dicercatalyzed step, might be <strong>in</strong>volved <strong>in</strong> <strong>the</strong> generation <strong>and</strong>function of at least some mi<strong>RNA</strong>s <strong>and</strong> si<strong>RNA</strong>s [3]. This issupported by <strong>the</strong> requirement of different Argonauteprote<strong>in</strong>s for <strong>the</strong> production of functional st<strong>RNA</strong>s or si<strong>RNA</strong>s<strong>in</strong> C. elegans [16,19,74]. In organisms where Dicer isencoded by a multigene family, such as Arabidopsis [57,58],cytoplasmic <strong>and</strong> nuclear process<strong>in</strong>g pathways mightoperate.Recent f<strong>in</strong>d<strong>in</strong>gs have also implicated small <strong>RNA</strong>s <strong>in</strong>chromat<strong>in</strong> <strong>and</strong>/or DNA modifications <strong>and</strong> genomerearrangements, such as heterochromat<strong>in</strong> formation<strong>in</strong> S. pombe <strong>and</strong> DNA elim<strong>in</strong>ation <strong>in</strong> Tetrahymena[1,35,49,65–68]. This suggests that ds<strong>RNA</strong>-mediatedprocesses might have a role <strong>in</strong> genome organization <strong>and</strong>transcriptional control. It is clear that despite muchprogress result<strong>in</strong>g from a comb<strong>in</strong>ation of genetics <strong>and</strong>biochemistry, we are only just beg<strong>in</strong>n<strong>in</strong>g to underst<strong>and</strong> <strong>the</strong>mechanistic complexity of <strong>RNA</strong>-mediated silenc<strong>in</strong>g, itsbiological implications, <strong>and</strong> <strong>the</strong> differences <strong>and</strong> similaritiesamong different eukaryotes.AcknowledgementsI thank S. Jacobsen, T. Clemente <strong>and</strong> members of mylaboratory for helpful comments. My apologies to colleagueswhose work could not be cited due to spacelimitations. Supported by NIH grant GM62915.References1 Matzke, M. et al. (2001) <strong>RNA</strong>: guid<strong>in</strong>g gene silenc<strong>in</strong>g. Science 293,1080–10832 Vance, V. <strong>and</strong> Vaucheret, H. (2001) <strong>RNA</strong> silenc<strong>in</strong>g <strong>in</strong> plants – defense<strong>and</strong> counterdefense. Science 292, 2277–22803 Hannon, G.J. (2002) <strong>RNA</strong> <strong><strong>in</strong>terference</strong>. Nature 418, 244–2514 Plasterk, R.H.A. (2002) <strong>RNA</strong> silenc<strong>in</strong>g: <strong>the</strong> genome’s immune system.Science 296, 1263–12655 Zamore, P.D. (2002) Ancient pathways programmed by small <strong>RNA</strong>s.Science 296, 1265–12696 Zamore, P.D. et al. (2000) <strong>RNA</strong>i: double-str<strong>and</strong>ed <strong>RNA</strong> directs <strong>the</strong>ATP-dependent cleavage of m<strong>RNA</strong> at 21 to 23 nucleotide <strong>in</strong>tervals. Cell101, 25–337 Bernste<strong>in</strong>, E. et al. (2001) Role for a bidentate ribonuclease <strong>in</strong> <strong>the</strong><strong>in</strong>itiation step of <strong>RNA</strong> <strong><strong>in</strong>terference</strong>. Nature 409, 363–3668 Hammond, S.M. et al. (2000) An <strong>RNA</strong>-directed nuclease mediates posttranscriptionalgene silenc<strong>in</strong>g <strong>in</strong> Drosophila <strong>cell</strong>s. Nature 404,293–2969 Nykänen, A. et al. (2001) ATP requirements <strong>and</strong> small <strong>in</strong>terfer<strong>in</strong>g <strong>RNA</strong>structure <strong>in</strong> <strong>the</strong> <strong>RNA</strong> <strong><strong>in</strong>terference</strong> pathway. Cell 107, 309–32110 Mart<strong>in</strong>ez, J. et al. (2002) S<strong>in</strong>gle-str<strong>and</strong>ed antisense si<strong>RNA</strong>s guidetarget <strong>RNA</strong> cleavage <strong>in</strong> <strong>RNA</strong>i. Cell 110, 563–57411 Elbashir, S.M. et al. (2001) <strong>RNA</strong> <strong><strong>in</strong>terference</strong> is mediated by 21- <strong>and</strong> 22-nucleotide <strong>RNA</strong>s. Genes Dev. 15, 188–20012 Holen, T. et al. (2002) Positional effects of short <strong>in</strong>terfer<strong>in</strong>g <strong>RNA</strong>starget<strong>in</strong>g <strong>the</strong> human coagulation trigger tissue factor. Nucleic AcidsRes. 30, 1757–176613 Han, Y. <strong>and</strong> Grierson, D. (2002) Relationship between small antisense<strong>RNA</strong>s <strong>and</strong> aberrant <strong>RNA</strong>s associated with sense transgene mediatedgene silenc<strong>in</strong>g <strong>in</strong> tomato. Plant J. 29, 509–51914 Hammond, S.M. et al. (2001) Argonaute2, a l<strong>in</strong>k between genetic <strong>and</strong>biochemical analyses of <strong>RNA</strong>i. Science 293, 1146–115015 Bohmert, K. et al. (1998) AGO1 def<strong>in</strong>es a novel locus of Arabidopsiscontroll<strong>in</strong>g leaf development. EMBO J. 17, 170–18016 Tabara, H. et al. (1999) The rde-1 gene, <strong>RNA</strong> <strong><strong>in</strong>terference</strong>, <strong>and</strong>transposon silenc<strong>in</strong>g <strong>in</strong> C. elegans. Cell 99, 123–13217 Hutvágner, G. <strong>and</strong> Zamore, P.D. (2002) A micro<strong>RNA</strong> <strong>in</strong> a multipleturnover<strong>RNA</strong>i enzyme complex. Science 297, 2056–206018 Cerutti, L. et al. (2000) Doma<strong>in</strong>s <strong>in</strong> gene silenc<strong>in</strong>g <strong>and</strong> <strong>cell</strong>differentiation prote<strong>in</strong>s: <strong>the</strong> novel PAZ doma<strong>in</strong> <strong>and</strong> redef<strong>in</strong>ition of<strong>the</strong> Piwi doma<strong>in</strong>. Trends Biochem. Sci. 25, 481–48219 Tabara, H. et al. (2002) The ds<strong>RNA</strong> b<strong>in</strong>d<strong>in</strong>g prote<strong>in</strong> RDE-4 <strong>in</strong>teractswith RDE-1, DCR-1, <strong>and</strong> a DExH-box helicase to direct <strong>RNA</strong>i <strong>in</strong>C. elegans. Cell 109, 861–87120 Grishok, A. et al. (2000) Genetic requirements for <strong>in</strong>heritance of <strong>RNA</strong>i<strong>in</strong> C. elegans. Science 287, 2494–249721 Kett<strong>in</strong>g, R.F. et al. (1999) Mut-7 of C. elegans, required for transposonsilenc<strong>in</strong>g <strong>and</strong> <strong>RNA</strong> <strong><strong>in</strong>terference</strong>, is a homolog of Werner syndromehelicase <strong>and</strong> RNaseD. Cell 99, 133–14122 Kennerdell, J.R. <strong>and</strong> Car<strong>the</strong>w, R.W. (1998) Use of ds<strong>RNA</strong>-mediatedgenetic <strong><strong>in</strong>terference</strong> to demonstrate that frizzled <strong>and</strong> frizzled 2 act <strong>in</strong><strong>the</strong> w<strong>in</strong>gless pathway. Cell 95, 1017–102623 Svoboda, P. et al. (2000) Selective reduction of dormant maternalm<strong>RNA</strong>s <strong>in</strong> mouse oocytes by <strong>RNA</strong> <strong><strong>in</strong>terference</strong>. Development 127,4147–415624 Ratcliff, F. et al. (1997) A similarity between viral defense <strong>and</strong> genesilenc<strong>in</strong>g <strong>in</strong> plants. Science 276, 1558–156025 Bitko, V. <strong>and</strong> Barik, S. (2001) Phenotypic silenc<strong>in</strong>g of cytoplasmicgenes us<strong>in</strong>g sequence-specific double-str<strong>and</strong>ed short <strong>in</strong>terfer<strong>in</strong>g <strong>RNA</strong><strong>and</strong> its application <strong>in</strong> <strong>the</strong> reverse genetics of wild type negative-str<strong>and</strong><strong>RNA</strong> viruses. BMC Microbiol. 1, 34–4526 Williams, R.W. <strong>and</strong> Rub<strong>in</strong>, G.M. (2002) ARGONAUTE1 is required forefficient <strong>RNA</strong> <strong><strong>in</strong>terference</strong> <strong>in</strong> Drosophila embryos. Proc. Natl Acad.Sci. USA 99, 6889–689427 Kataoka, Y. et al. (2001) Developmental roles <strong>and</strong> molecularcharacterization of a Drosophila homologue of Arabidopsis Argonaute1,<strong>the</strong> founder of a novel gene superfamily. Genes Cells 6,313–32528 Harris, A.N. <strong>and</strong> Macdonald, P.M. (2001) auberg<strong>in</strong>e encodes aDrosophila polar granule component required for pole <strong>cell</strong> formation<strong>and</strong> related to eIF2C. Development 128, 2823–283229 Zeng, Y. <strong>and</strong> Cullen, B.R. (2002) <strong>RNA</strong> <strong><strong>in</strong>terference</strong> <strong>in</strong> human <strong>cell</strong>s isrestricted to <strong>the</strong> cytoplasm. <strong>RNA</strong> 8, 855–86030 Billy, E. et al. (2001) Specific <strong><strong>in</strong>terference</strong> with gene expression <strong>in</strong>ducedby long, double-str<strong>and</strong>ed <strong>RNA</strong> <strong>in</strong> mouse embryonal teratocarc<strong>in</strong>oma<strong>cell</strong> l<strong>in</strong>es. Proc. Natl Acad. Sci. USA 98, 14428–1443331 Cikaluk, D.E. et al. (1999) GERp95, a membrane-associated prote<strong>in</strong>that belongs to a family of prote<strong>in</strong>s <strong>in</strong>volved <strong>in</strong> stem <strong>cell</strong> differentiation.Mol. Biol. Cell 10, 3357–337232 Wesley, S.V. et al. (2001) Construct design for efficient, effective <strong>and</strong>high-throughput gene silenc<strong>in</strong>g <strong>in</strong> plants. Plant J. 27, 581–59033 Kalidas, S. <strong>and</strong> Smith, D.P. (2002) Novel genomic cDNA hybridshttp://tigs.trends.com