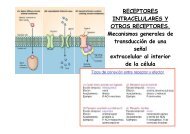
Cover Picture: Proteomics 9'11
Cover Picture: Proteomics 9'11
Cover Picture: Proteomics 9'11
- No tags were found...
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
1562 G. K. Agrawal et al. <strong>Proteomics</strong> 2011, 11, 1559–1568functions and pathways (e.g. databases based on Geneontologies, MAPMAN and PPDB) [68–70], and wouldbenefit from links with transcriptome and metabolome data.It is worthwhile noting that proteomics can be instrumentalin linking quantitative genetics and physiology, in particularby mapping PQLs (protein QTLs (quantitative trait loci),[71–73]). While eQTLs (expression QTLs) have been detectedby using transcriptome microarrays, the link with metabolismis relatively loose, partly because of PTMs, includingthose involved in protein turn over. Metabolomics profilingprovides very interesting phenotypic data and metaboliteQTLs can be mapped [74]. The integration of metabolomicswith transcriptomics and proteomics is another innovativeway to assign gene functions within a genome and analyzeplant responses at a systems level [33, 75].As proteins are the final products of genes, they potentiallyaccount better than transcripts for activity variationssimply because: (i) their accumulation level is the result of acombination of transcription, translation and protein turnover,and (ii) proteomics studies can also analyze PTMs thatmodulate their activity. All causes of these variations canpotentially be studied, from genes regulating transcriptionto enzymes responsible for PTMs. PTMs affect enzymeactivity and subcellular localization, participate in signaling(e.g. phosphorylation) and play important roles in thematuration of storage proteins. Most of these phenomenacannot be predicted from genome or transcriptome data. Afeature of most plants living in temperate climates is theirreliance on dehydrated stages including seeds [76] andpollen grains [77, 78], where the regulation of mRNA andprotein abundances are particularly disconnected and wherePTMs play an important role.The throughput of proteomics studies is still lower thanthat of transcriptomics studies regarding the number ofgenes and proteins analyzed. The number of proteins thatcan be identified and quantified in a single shotgun MS/MSbasedproteomics experiment using current technology istypically from 500 to several thousands at maximum. Incontrast, tens of thousands of predicted transcripts of anorganism can be analyzed in one experiment (microarrays,SAGE or new generation RNA sequencing), includingquantification of transcript expression levels. However,because cellular reactions directly depend on their amountand state, the analysis of protein variation is potentiallymore informative than the analysis of transcript abundancesfor the analysis of the determinism of quantitative traits.Experimental designs for this type of analysis comprisenumerous genotypes (e.g. populations of recombinantinbred lines). Although up to 200–300 2-D gels can inprinciple be analyzed in the same experiment (e.g. [71, 79]),this is not practical for most laboratories due to the highcosts of materials and labor, and complex tracking andimage analysis. Advances in MS [2], and particularly in labelfreequantification, have made possible the analysis of verylarge data sets in complex multi-variate experimentaldesigns [33, 80]. These approaches will allow the detection ofprotein QTLs and the quantification of candidate proteins inlarge panels for association genetics, enabling new ‘associationproteomics’ programs in plants. Last but not least,continuous progress in sub-cellular identification of proteinsallows pushing the limits toward a detailed understandingof plant proteomes at the sub-organelle, organelle, cell,tissue and organ levels [58, 81].3 The rationale and birth of InternationalPlant <strong>Proteomics</strong> Organization (INPPO)Nevertheless, there is something lacking in our questtoward the ambitious goal of achieving complete understandingof proteomes of plant species. There are manyreasons that can be proposed to explain this gap in ourknowledge. To suggest a few, there is lack of access toproteomics in most plant laboratories around the world, lackof information, lack of close cooperation among proteomicsresearchers and most importantly lack of a suitable platformto globally discuss plant proteomics from both fundamentaland applied perspectives. In addition, there is a morepressing concern that remains to be addressed, which is theincredibly large diversity of plants and crops on our planet,which means different things to different people andsocieties.Only a very few plants are the subject of intensiveresearch efforts, the vast majority are not. For example,Arabidopsis [82, 83] is studied as a model plant largelybecause it was the first plant to be sequenced, has wellestablishedgene manipulation techniques [84], a quick lifecycle and is very easy to handle. Rice, on the other hand, hasa relatively long life cycle, and is difficult to handle, but isbeing investigated as a model for cereal monocot crops(such as wheat, rye, barley and maize), largely because it isthe primary food source of half the world’s population.Oilseeds, other than providing a rich source of energy in ourfood, are being economically exploited as bio-fuel producers.The same holds true for sugarcane and sugarbeet for thesugar they provide us, grapes for wine production andcotton in textile industries. Poplar has been used as themodel plant for woody species [85], for economic and ecophysiologicalreasons, as e.g. it has a wide distribution in thenorthern hemisphere, and hence can be transformed andeasily in vitro propagated. The poplar model has been usedin many studies to provide data on the effects of environmentalstressors in woody plans, and to investigate featuresspecific to woody species, i.e. formation of secondary tissues,overwintering, etc. [86]. Figure 3 highlights some of themajor plants/crops in the world along with their geographicaldistribution. Looking globally, one thing stands out –‘global impact of global plant proteomics.’ In other words,the importance of proteomics in the biological context, plantdiversity and the essential contribution of that biodiversity tothe socio-economic fabric of humankind is beyond anydoubt. Although we would like to claim that proteomics of& 2011 WILEY-VCH Verlag GmbH & Co. KGaA, Weinheimwww.proteomics-journal.com