Guidelines and Handbook for IBSCs - Department of Biotechnology
Guidelines and Handbook for IBSCs - Department of Biotechnology
Guidelines and Handbook for IBSCs - Department of Biotechnology
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
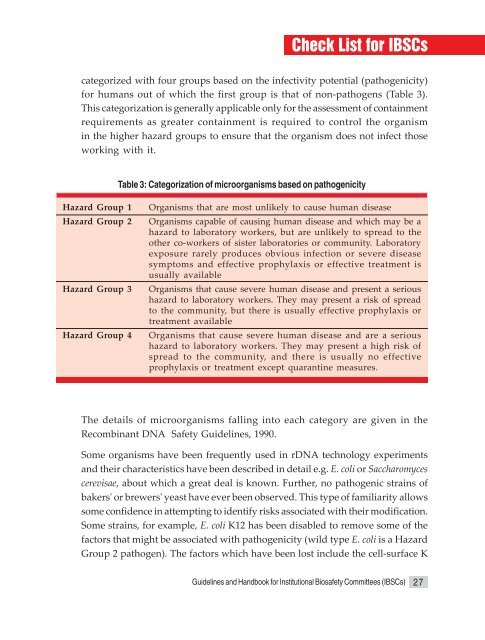
Check List <strong>for</strong> <strong>IBSCs</strong>categorized with four groups based on the infectivity potential (pathogenicity)<strong>for</strong> humans out <strong>of</strong> which the first group is that <strong>of</strong> non-pathogens (Table 3).This categorization is generally applicable only <strong>for</strong> the assessment <strong>of</strong> containmentrequirements as greater containment is required to control the organismin the higher hazard groups to ensure that the organism does not infect thoseworking with it.Table 3: Categorization <strong>of</strong> microorganisms based on pathogenicityHazard Group 1Hazard Group 2Hazard Group 3Hazard Group 4Organisms that are most unlikely to cause human diseaseOrganisms capable <strong>of</strong> causing human disease <strong>and</strong> which may be ahazard to laboratory workers, but are unlikely to spread to theother co-workers <strong>of</strong> sister laboratories or community. Laboratoryexposure rarely produces obvious infection or severe diseasesymptoms <strong>and</strong> effective prophylaxis or effective treatment isusually availableOrganisms that cause severe human disease <strong>and</strong> present a serioushazard to laboratory workers. They may present a risk <strong>of</strong> spreadto the community, but there is usually effective prophylaxis ortreatment availableOrganisms that cause severe human disease <strong>and</strong> are a serioushazard to laboratory workers. They may present a high risk <strong>of</strong>spread to the community, <strong>and</strong> there is usually no effectiveprophylaxis or treatment except quarantine measures.The details <strong>of</strong> microorganisms falling into each category are given in theRecombinant DNA Safety <strong>Guidelines</strong>, 1990.Some organisms have been frequently used in rDNA technology experiments<strong>and</strong> their characteristics have been described in detail e.g. E. coli or Saccharomycescerevisae, about which a great deal is known. Further, no pathogenic strains <strong>of</strong>bakers' or brewers' yeast have ever been observed. This type <strong>of</strong> familiarity allowssome confidence in attempting to identify risks associated with their modification.Some strains, <strong>for</strong> example, E. coli K12 has been disabled to remove some <strong>of</strong> thefactors that might be associated with pathogenicity (wild type E. coli is a HazardGroup 2 pathogen). The factors which have been lost include the cell-surface K<strong>Guidelines</strong> <strong>and</strong> H<strong>and</strong>book <strong>for</strong> Institutional Biosafety Committees (<strong>IBSCs</strong>) 27