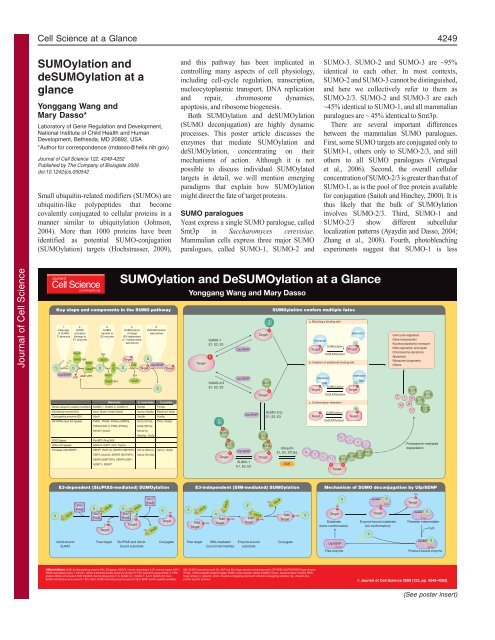
SUMOylation and DeSUMOylation at a Glance - Journal of Cell ...
SUMOylation and DeSUMOylation at a Glance - Journal of Cell ...
SUMOylation and DeSUMOylation at a Glance - Journal of Cell ...
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
<strong>Journal</strong> <strong>of</strong> <strong>Cell</strong> Science<br />
<strong>Cell</strong> Science <strong>at</strong> a <strong>Glance</strong><br />
<strong>SUMOyl<strong>at</strong>ion</strong> <strong>and</strong><br />
de<strong>SUMOyl<strong>at</strong>ion</strong> <strong>at</strong> a<br />
glance<br />
Yonggang Wang <strong>and</strong><br />
Mary Dasso*<br />
Labor<strong>at</strong>ory <strong>of</strong> Gene Regul<strong>at</strong>ion <strong>and</strong> Development,<br />
N<strong>at</strong>ional Institute <strong>of</strong> Child Health <strong>and</strong> Human<br />
Development, Bethesda, MD 20892, USA<br />
*Author for correspondence (mdasso@helix.nih.gov)<br />
<strong>Journal</strong> <strong>of</strong> <strong>Cell</strong> Science 122, 4249-4252<br />
Published by The Company <strong>of</strong> Biologists 2009<br />
doi:10.1242/jcs.050542<br />
Small ubiquitin-rel<strong>at</strong>ed modifiers (SUMOs) are<br />
ubiquitin-like polypeptides th<strong>at</strong> become<br />
covalently conjug<strong>at</strong>ed to cellular proteins in a<br />
manner similar to ubiquityl<strong>at</strong>ion (Johnson,<br />
2004). More than 1000 proteins have been<br />
identified as potential SUMO-conjug<strong>at</strong>ion<br />
(<strong>SUMOyl<strong>at</strong>ion</strong>) targets (Hochstrasser, 2009),<br />
jcs.biologists.org<br />
Key steps <strong>and</strong> components in the SUMO p<strong>at</strong>hway<br />
1.<br />
2.<br />
Cleavage SUMO<br />
<strong>of</strong> SUMO activ<strong>at</strong>ion<br />
C-terminus (linkage to<br />
E1 enzyme)<br />
Aos1<br />
E3s Ulp/SENP<br />
Ulp/SENP<br />
ATP<br />
SH<br />
AMP+PPi<br />
Aos1<br />
SH<br />
Uba2 SH Ubc9<br />
K<br />
Target S<br />
S S Uba2 S S S S<br />
K<br />
Target<br />
Aos1<br />
Uba2 SH Ubc9<br />
Abbrevi<strong>at</strong>ions: DUB, deubiquityl<strong>at</strong>ing enzyme; E3s, E3 ligases; HDAC4, histone deacetylase 4; IR, internal repe<strong>at</strong>; KAP1,<br />
KRAB-associ<strong>at</strong>ed protein 1; Mms21, methyl methanesulfon<strong>at</strong>e sensitivity protein 21; Pc2, polycomb group protein 2; PIAS,<br />
protein inhibitor <strong>of</strong> activ<strong>at</strong>ed STAT; RanBP2, Ran binding protein 2; S, SUMO; S-1, SUMO-1; S-2/3, SUMO-2/3; Sae1,<br />
SUMO-activ<strong>at</strong>ing enzyme subunit 1 (E1); Sae2, SUMO-activ<strong>at</strong>ing enzyme subunit 2 (E1); SENP, sentrin-specific protease;<br />
<strong>and</strong> this p<strong>at</strong>hway has been implic<strong>at</strong>ed in<br />
controlling many aspects <strong>of</strong> cell physiology,<br />
including cell-cycle regul<strong>at</strong>ion, transcription,<br />
nucleocytoplasmic transport, DNA replic<strong>at</strong>ion<br />
<strong>and</strong> repair, chromosome dynamics,<br />
apoptosis, <strong>and</strong> ribosome biogenesis.<br />
Both <strong>SUMOyl<strong>at</strong>ion</strong> <strong>and</strong> de<strong>SUMOyl<strong>at</strong>ion</strong><br />
(SUMO deconjug<strong>at</strong>ion) are highly dynamic<br />
processes. This poster article discusses the<br />
enzymes th<strong>at</strong> medi<strong>at</strong>e <strong>SUMOyl<strong>at</strong>ion</strong> <strong>and</strong><br />
de<strong>SUMOyl<strong>at</strong>ion</strong>, concentr<strong>at</strong>ing on their<br />
mechanisms <strong>of</strong> action. Although it is not<br />
possible to discuss individual SUMOyl<strong>at</strong>ed<br />
targets in detail, we will mention emerging<br />
paradigms th<strong>at</strong> explain how <strong>SUMOyl<strong>at</strong>ion</strong><br />
might direct the f<strong>at</strong>e <strong>of</strong> target proteins.<br />
SUMO paralogues<br />
Yeast express a single SUMO paralogue, called<br />
Smt3p in Saccharomyces cerevisiae.<br />
Mammalian cells express three major SUMO<br />
paralogues, called SUMO-1, SUMO-2 <strong>and</strong><br />
<strong>SUMOyl<strong>at</strong>ion</strong> <strong>and</strong> De<strong>SUMOyl<strong>at</strong>ion</strong> <strong>at</strong> a <strong>Glance</strong><br />
4.<br />
<strong>SUMOyl<strong>at</strong>ion</strong><br />
<strong>of</strong> target<br />
(E3-dependent<br />
or -independent;<br />
see below)<br />
K<br />
Target<br />
Mammals S. cerevisiae S. pombe<br />
Small ubiquitin-rel<strong>at</strong>ed modifiers SUMO-1, SUMO-2, SUMO-3 Smt3p Pmt3p<br />
Activ<strong>at</strong>ing enzyme (E1)<br />
Aos1 (Sae1)-Uba2 (Sae2)<br />
Aos1p-Uba2p Rad31p-Fub2p<br />
Conjug<strong>at</strong>ing enzyme (E2) Ubc9<br />
Ubc9p Hus5p<br />
SP-RING-type E3 ligases PIAS1, PIAS3, PIASxα (ARIP3), Siz1p (UII1p), Pli1p, Nse2p<br />
PIASxβ (Miz1), PIASγ (PIAS4), Siz2p (Nfi1p),<br />
Mms21 (Nse2)<br />
Mms21p<br />
(Nse2p), Zip3p<br />
IR E3 ligase<br />
Other E3 ligases<br />
Protease (Ulp/SENP)<br />
3.<br />
SUMO<br />
transfer to<br />
E2 enzyme<br />
3.<br />
De<strong>SUMOyl<strong>at</strong>ion</strong><br />
(see below)<br />
RanBP2 (Nup358)<br />
HDAC4, KAP1, Pc2, Topors<br />
SENP1 (SuPr-2), SENP2 (SMT3IP2, UIp1p (Nib1p), Ulp1p, Ulp2p<br />
SSP3, Axam2), SENP3 (SMT3IP1), Ulp2p (Smt4p)<br />
SENP5 (SMT3IP3), SENP6 (SSP1,<br />
SUSP1), SENP7<br />
S<br />
Yonggang Wang <strong>and</strong> Mary Dasso<br />
Target<br />
SUMO-1<br />
E1, E2, E3<br />
K<br />
Ulp/SENP<br />
Ulp/SENP<br />
Ulp/SENP<br />
<strong>SUMOyl<strong>at</strong>ion</strong> confers multiple f<strong>at</strong>es<br />
Ubiquitin<br />
E1, E2, STUbL<br />
SIM, SUMO-interacting motif; Siz, SAP <strong>and</strong> Miz-finger domain-containing protein; SP-RING, Siz/PIAS RING-finger domain;<br />
STUbL, SUMO-targeted ubiquitin ligase; SUMO, small ubiquitin-rel<strong>at</strong>ed modifier; Topors, topoisomerase I-binding RING<br />
finger protein; U, ubiquitin; Ubc9, ubiquitin-conjug<strong>at</strong>ing enzyme E2 I (SUMO-conjug<strong>at</strong>ing enzyme); Ulp, ubiquitin-likeprotein-specific<br />
protease.<br />
4249<br />
SUMO-3. SUMO-2 <strong>and</strong> SUMO-3 are ~95%<br />
identical to each other. In most contexts,<br />
SUMO-2 <strong>and</strong> SUMO-3 cannot be distinguished,<br />
<strong>and</strong> here we collectively refer to them as<br />
SUMO-2/3. SUMO-2 <strong>and</strong> SUMO-3 are each<br />
~45% identical to SUMO-1, <strong>and</strong> all mammalian<br />
paralogues are ~ 45% identical to Smt3p.<br />
There are several important differences<br />
between the mammalian SUMO paralogues.<br />
First, some SUMO targets are conjug<strong>at</strong>ed only to<br />
SUMO-1, others only to SUMO-2/3, <strong>and</strong> still<br />
others to all SUMO paralogues (Vertegaal<br />
et al., 2006). Second, the overall cellular<br />
concentr<strong>at</strong>ion <strong>of</strong> SUMO-2/3 is gre<strong>at</strong>er than th<strong>at</strong> <strong>of</strong><br />
SUMO-1, as is the pool <strong>of</strong> free protein available<br />
for conjug<strong>at</strong>ion (Saitoh <strong>and</strong> Hinchey, 2000). It is<br />
thus likely th<strong>at</strong> the bulk <strong>of</strong> <strong>SUMOyl<strong>at</strong>ion</strong><br />
involves SUMO-2/3. Third, SUMO-1 <strong>and</strong><br />
SUMO-2/3 show different subcellular<br />
localiz<strong>at</strong>ion p<strong>at</strong>terns (Ayaydin <strong>and</strong> Dasso, 2004;<br />
Zhang et al., 2008). Fourth, photobleaching<br />
experiments suggest th<strong>at</strong> SUMO-1 is less<br />
E3-dependent (Siz/PIAS-medi<strong>at</strong>ed) <strong>SUMOyl<strong>at</strong>ion</strong> E3-independent (SIM-medi<strong>at</strong>ed) <strong>SUMOyl<strong>at</strong>ion</strong><br />
Mechanism <strong>of</strong> SUMO deconjug<strong>at</strong>ion by Ulp/SENP<br />
O<br />
Gly<br />
S S<br />
Cys<br />
Ubc9<br />
Ubc9-bound<br />
SUMO<br />
Siz/<br />
PIAS<br />
Ubc9<br />
Ubc9<br />
Ubc9<br />
Free target Siz/PIAS <strong>and</strong> Ubc9bound<br />
substr<strong>at</strong>e<br />
Ubc9<br />
Ubc9<br />
SUMO-2/3<br />
E1, E2, E3<br />
Conjug<strong>at</strong>e Free target SIM-medi<strong>at</strong>ed<br />
bound intermedi<strong>at</strong>e<br />
S<br />
S-1<br />
K<br />
S-2/3<br />
K<br />
S-2/3n<br />
Ulp/SENP<br />
K<br />
Target<br />
SUMO-1<br />
E1, E2, E3<br />
O<br />
Gly<br />
S<br />
Cys<br />
Enzyme-bound<br />
substr<strong>at</strong>e<br />
S-2/3<br />
K<br />
Target<br />
SUMO-2/3n<br />
E1, E2, E3<br />
S-2/3<br />
K<br />
S-2/3n<br />
K<br />
Target<br />
Siz/<br />
O<br />
O<br />
PIAS<br />
O<br />
Gly<br />
S S<br />
Gly<br />
Ubc9<br />
S S<br />
Gly O<br />
Gly<br />
Cys<br />
Cys<br />
S<br />
S S<br />
NH<br />
Cys<br />
Siz/<br />
Siz/<br />
NH2<br />
K<br />
PIAS<br />
PIAS<br />
K<br />
SIM K NH2<br />
NH2<br />
Target<br />
SIM K NH2 Target<br />
K<br />
Target<br />
Target<br />
Target<br />
HS<br />
Cys<br />
SIM<br />
Target<br />
K NH2<br />
O<br />
SIM K NH<br />
Target<br />
S<br />
S<br />
Gly<br />
S<br />
Cys<br />
O<br />
Ubc9<br />
S-1<br />
K<br />
Target<br />
HS<br />
Cys<br />
Ubc9<br />
DUB<br />
Conjug<strong>at</strong>e<br />
Gly<br />
Un<br />
a. Blocking a binding site<br />
Interactor<br />
K<br />
Target arget<br />
<strong>SUMOyl<strong>at</strong>ion</strong><br />
S<br />
K<br />
Target Ta T rget<br />
U<br />
U<br />
De<strong>SUMOyl<strong>at</strong>ion</strong><br />
b. Cre<strong>at</strong>ion <strong>of</strong> additional binding site<br />
Interactor<br />
Interactor<br />
SIM<br />
SIM<br />
S<br />
K <strong>SUMOyl<strong>at</strong>ion</strong><br />
K<br />
Target<br />
Target<br />
De<strong>SUMOyl<strong>at</strong>ion</strong><br />
c. Conform<strong>at</strong>ion alter<strong>at</strong>ion<br />
K <strong>SUMOyl<strong>at</strong>ion</strong><br />
Target<br />
De<strong>SUMOyl<strong>at</strong>ion</strong><br />
Un<br />
S-2/3<br />
U S-2/3<br />
S-2/3<br />
S-2/3n<br />
K K<br />
Target<br />
S<br />
Gly<br />
O<br />
NH<br />
K<br />
Target<br />
Substr<strong>at</strong>e<br />
(trans conform<strong>at</strong>ion)<br />
Ulp/SENP<br />
HS-Cys<br />
Free enzyme<br />
S<br />
K<br />
Target<br />
U<br />
Interactor<br />
U<br />
U<br />
S<br />
O Gly<br />
OH<br />
Ulp/SENP S<br />
O Gly<br />
S-Cys<br />
NH<br />
K<br />
Target<br />
Enzyme-bound substr<strong>at</strong>e<br />
(cis conform<strong>at</strong>ion)<br />
<strong>Cell</strong>-cycle regul<strong>at</strong>ion<br />
Gene transcription<br />
Nucleocytoplasmic transport<br />
DNA replic<strong>at</strong>ion <strong>and</strong> repair<br />
Chromosome dynamics<br />
Apoptosis<br />
Ribosome biogenesis<br />
Others<br />
S-2/3<br />
S-2/3<br />
T r e<br />
S-2/3<br />
a<br />
g<br />
t<br />
S-2/3n<br />
Proteasome-medi<strong>at</strong>ed<br />
degrad<strong>at</strong>ion<br />
NH2<br />
K<br />
Target<br />
Ulp/SENP S<br />
O Gly<br />
S-Cys<br />
Thioester intermedi<strong>at</strong>e<br />
H2O<br />
Ulp/SENP S<br />
O Gly<br />
OH HS-Cys<br />
Product-bound enzyme<br />
© <strong>Journal</strong> <strong>of</strong> <strong>Cell</strong> Science 2009 (122, pp. 4249-4252)<br />
(See poster insert)
<strong>Journal</strong> <strong>of</strong> <strong>Cell</strong> Science<br />
4250<br />
<strong>Journal</strong> <strong>of</strong> <strong>Cell</strong> Science 122 (23)<br />
dynamic than the other SUMO paralogues, <strong>and</strong><br />
its p<strong>at</strong>tern <strong>of</strong> conjug<strong>at</strong>ion responds differently to<br />
he<strong>at</strong> shock <strong>and</strong> stress (Ayaydin <strong>and</strong> Dasso, 2004;<br />
Saitoh <strong>and</strong> Hinchey, 2000).<br />
Smt3p, SUMO-2 <strong>and</strong> SUMO-3 can form<br />
conjug<strong>at</strong>ed chains through a single conserved<br />
acceptor lysine (Bylebyl et al., 2003; T<strong>at</strong>ham<br />
et al., 2001). SUMO-1 does not have an<br />
equivalent lysine residue, <strong>and</strong> thus probably<br />
does not act as a link in elong<strong>at</strong>ing chains<br />
in vivo. However, SUMO-1 might termin<strong>at</strong>e<br />
chains th<strong>at</strong> are elong<strong>at</strong>ed through serial<br />
conjug<strong>at</strong>ion <strong>of</strong> SUMO-2/3 (M<strong>at</strong>ic et al., 2008).<br />
Notably, although <strong>SUMOyl<strong>at</strong>ion</strong> does not seem<br />
to rely upon the geometry <strong>of</strong> chain linkages to<br />
confer inform<strong>at</strong>ion, as ubiquityl<strong>at</strong>ion does<br />
(Pickart <strong>and</strong> Fushman, 2004), SUMO<br />
conjug<strong>at</strong>es built from different paralogues or<br />
with different chain lengths can specify distinct<br />
target f<strong>at</strong>es, as discussed below.<br />
SUMO-interacting motifs<br />
SUMO-interacting motifs (SIMs) play a central<br />
role in both the enzymology <strong>of</strong> the SUMO<br />
p<strong>at</strong>hway <strong>and</strong> in the f<strong>at</strong>e <strong>of</strong> conjug<strong>at</strong>ed species.<br />
The best-characterized class <strong>of</strong> SIM consists <strong>of</strong> a<br />
hydrophobic core ([V/I]-x-[V/I]-[V/I]) flanked<br />
by a cluster <strong>of</strong> neg<strong>at</strong>ively charged amino acids<br />
(Kerscher, 2007). The SIM hydrophobic core<br />
can bind to an interaction surface on SUMO<br />
proteins in a parallel or antiparallel orient<strong>at</strong>ion.<br />
The acidic residues adjacent to the core might<br />
contribute to the affinity, orient<strong>at</strong>ion or<br />
paralogue specificity <strong>of</strong> binding (Hecker et al.,<br />
2006; Meulmeester et al., 2008). A variant SIM<br />
was recently defined within the transcriptional<br />
repressor CoREST1, consisting <strong>of</strong> [I/V/L]-<br />
[D/E]-[I/V/L]-[D/E]-[I/V/L] with N-terminal<br />
acidic residues (Ouyang et al., 2009). This SIM<br />
is highly selective for SUMO-2/3 binding, <strong>and</strong><br />
differs from previously identified SIMs because<br />
its core lacks a hydrophobic residue <strong>at</strong> position<br />
4. Notably, the diversity <strong>of</strong> SIMs identified to<br />
d<strong>at</strong>e is much less than the 16 known ubiquitinbinding<br />
domains (Grabbe <strong>and</strong> Dikic, 2009); it is<br />
reasonable to specul<strong>at</strong>e th<strong>at</strong> additional SIMs<br />
remain to be discovered.<br />
SUMO-processing, -activ<strong>at</strong>ing <strong>and</strong><br />
-conjug<strong>at</strong>ing enzymes<br />
Newly transl<strong>at</strong>ed SUMO proteins are cleaved to<br />
reveal C-terminal diglycine motifs. This<br />
processing is medi<strong>at</strong>ed by a family <strong>of</strong><br />
proteases known as ubiquitin-like-proteinspecific<br />
proteases (Ulps) in yeast <strong>and</strong><br />
sentrin-specific proteases (SENPs) in mammals<br />
(Mukhopadhyay <strong>and</strong> Dasso, 2007). Ulps <strong>and</strong><br />
SENPs also medi<strong>at</strong>e de<strong>SUMOyl<strong>at</strong>ion</strong> (see<br />
below).<br />
All SUMO paralogues share the same<br />
activ<strong>at</strong>ing (E1) <strong>and</strong> conjug<strong>at</strong>ing (E2) enzymes.<br />
These enzymes are structurally similar to E1 <strong>and</strong><br />
E2 enzymes <strong>of</strong> ubiquitin, <strong>and</strong> they share many <strong>of</strong><br />
the properties th<strong>at</strong> have been demonstr<strong>at</strong>ed for<br />
those enzymes (Hochstrasser, 2009). The<br />
yeast SUMO E1 enzyme is a heterodimer<br />
consisting <strong>of</strong> Aos1p (also known as Sae1 in<br />
vertebr<strong>at</strong>es) <strong>and</strong> Uba2p (also known as Sae2<br />
in vertebr<strong>at</strong>es), which show sequence similarity<br />
to the N-terminus <strong>and</strong> C-terminus <strong>of</strong> the<br />
monomeric ubiquitin E1 enzyme, respectively<br />
(Johnson et al., 1997). Aos1p-Uba2p c<strong>at</strong>alyzes<br />
the form<strong>at</strong>ion <strong>of</strong> a high-energy thioester bond<br />
between Uba2p <strong>and</strong> the SUMO C-terminus,<br />
with ATP hydrolysis to AMP <strong>and</strong> pyrophosph<strong>at</strong>e<br />
(Johnson et al., 1997; Lois <strong>and</strong> Lima, 2005). The<br />
activ<strong>at</strong>ed SUMO is subsequently passed to a<br />
cysteine in the active site <strong>of</strong> the E2 enzyme,<br />
Ubc9, through an intermolecular thiol-transfer<br />
reaction.<br />
Residues <strong>of</strong> Ubc9 th<strong>at</strong> are directly involved in<br />
the transfer <strong>of</strong> SUMO act to orient the lysine<br />
<strong>of</strong> the target protein <strong>and</strong> to decrease its pKa,<br />
resulting in a higher occurrence <strong>of</strong> its activ<strong>at</strong>ed,<br />
de-proton<strong>at</strong>ed st<strong>at</strong>e (Yunus <strong>and</strong> Lima, 2006).<br />
SUMO transfer from Ubc9 to some target<br />
proteins can occur through <strong>at</strong> least two ligaseindependent<br />
mechanisms. First, many<br />
SUMOyl<strong>at</strong>ed lysines lie within a consensus<br />
motif, �-K-x-[D/E] (where � is an aliph<strong>at</strong>ic<br />
branched amino acid <strong>and</strong> x is any amino acid).<br />
Ubc9 can directly recognize this motif <strong>and</strong><br />
conjug<strong>at</strong>e the lysine residue within it (Bernier-<br />
Villamor et al., 2002). Second, some SUMO<br />
substr<strong>at</strong>es contain SIMs th<strong>at</strong> promote their own<br />
conjug<strong>at</strong>ion (Meulmeester et al., 2008; Zhu<br />
et al., 2008). These SIMs bind to the SUMO<br />
moiety to which Ubc9 is <strong>at</strong>tached, thereby<br />
increasing its local concentr<strong>at</strong>ion <strong>and</strong><br />
facilit<strong>at</strong>ing <strong>SUMOyl<strong>at</strong>ion</strong>. Because SIMs show<br />
paralogue preference, this mechanism allows<br />
targets to be modified in a paralogue-selective<br />
manner. Notably, mammalian Ubc9 can itself be<br />
SUMOyl<strong>at</strong>ed on a nonconsensus lysine in its<br />
N-terminal helix (Knipscheer et al., 2008). This<br />
modific<strong>at</strong>ion does not inhibit its activity per se,<br />
but alters its target preference, increasing the<br />
conjug<strong>at</strong>ion <strong>of</strong> substr<strong>at</strong>es th<strong>at</strong> contain SIMs,<br />
which bind to the SUMO th<strong>at</strong> is conjug<strong>at</strong>ed to<br />
Ubc9.<br />
SUMO ligases<br />
SUMO ligases (E3 enzymes) facilit<strong>at</strong>e the<br />
majority <strong>of</strong> <strong>SUMOyl<strong>at</strong>ion</strong> under physiological<br />
conditions (Meulmeester et al., 2008). A number<br />
<strong>of</strong> SUMO ligases have been described, most <strong>of</strong><br />
which seem to be specific to metazoans.<br />
Siz/PIAS-family proteins<br />
All eukaryotes express proteins with Siz/PIAS<br />
RING-finger-like domains (SP-RING<br />
domains), which are known as SAP <strong>and</strong> Miz-<br />
finger domain (Siz) proteins in yeast <strong>and</strong><br />
protein inhibitor <strong>of</strong> activ<strong>at</strong>ed STAT (PIAS)<br />
proteins in vertebr<strong>at</strong>es (Hochstrasser, 2001). In<br />
budding yeast, Siz1p <strong>and</strong> Siz2p are required for<br />
most Smt3p conjug<strong>at</strong>ion (Johnson <strong>and</strong> Gupta,<br />
2001; Takahashi et al., 2001). Other SP-RING<br />
proteins, Zip3p <strong>and</strong> Mms21p, promote<br />
assembly <strong>of</strong> the synaptonemal complex<br />
between homologous chromosomes during<br />
meiosis (Cheng et al., 2006) <strong>and</strong> DNA repair<br />
(Potts, 2009), respectively. The five vertebr<strong>at</strong>e<br />
PIAS proteins (PIAS1, PIAS3, PIASx�,<br />
PIASx� <strong>and</strong> PIAS�) have been implic<strong>at</strong>ed in<br />
many processes, including gene expression,<br />
signal transduction <strong>and</strong> genome maintenance<br />
(Palvimo, 2007).<br />
Beyond their SP-RING domains, Siz/PIASfamily<br />
members share additional conserved<br />
motifs, including an N-terminal scaffold<br />
<strong>at</strong>tachment factor (SAF)-A/B, acinus, PIAS<br />
(SAP) motif, a PINIT motif, a SIM, <strong>and</strong> a<br />
C-terminal domain th<strong>at</strong> is rich in serine <strong>and</strong><br />
acidic amino acids (S/DE domain) (Palvimo,<br />
2007). The SAP domain directs the localiz<strong>at</strong>ion<br />
<strong>of</strong> Siz/PIAS proteins to chrom<strong>at</strong>in within the<br />
nucleus (Azuma et al., 2005; Palvimo, 2007).<br />
Structural analysis <strong>of</strong> a Siz1 fragment th<strong>at</strong> is<br />
sufficient for E3 activity in vitro shows th<strong>at</strong> it<br />
has an elong<strong>at</strong>ed tripartite architecture, formed<br />
by its N-terminal PINIT domain, SP-RING<br />
domain <strong>and</strong> C-terminal domain, termed the<br />
SP-CTD (Yunus <strong>and</strong> Lima, 2009). The SP-<br />
RING <strong>and</strong> SP-CTD domains are required for<br />
activ<strong>at</strong>ion <strong>of</strong> the Ubc9-SUMO thioester,<br />
whereas the PINIT domain directs<br />
<strong>SUMOyl<strong>at</strong>ion</strong> to the correct target lysine.<br />
RanBP2<br />
RanBP2 is a nuclear-pore protein th<strong>at</strong> localizes<br />
to the cytoplasmic face <strong>of</strong> the pore. RanBP2<br />
possesses a domain called the internal repe<strong>at</strong><br />
(IR) domain, which consists <strong>of</strong> two t<strong>and</strong>emly<br />
repe<strong>at</strong>ed sequences <strong>of</strong> around 50 residues (IR1<br />
<strong>and</strong> IR2), separ<strong>at</strong>ed by a 24-residue spacer (M).<br />
RanBP2 fragments containing the IR domain<br />
have SUMO ligase activity in vitro (Pichler<br />
et al., 2002). Structural analysis <strong>of</strong> a RanBP2<br />
fragment containing the IR1 <strong>and</strong> M domains<br />
indic<strong>at</strong>es th<strong>at</strong> RanBP2 enhances Ubc9 activity<br />
without direct contacts to the target protein<br />
(Reverter <strong>and</strong> Lima, 2005). It has thus been<br />
proposed th<strong>at</strong> RanBP2 promotes <strong>SUMOyl<strong>at</strong>ion</strong><br />
by aligning the Ubc9-SUMO thioester complex<br />
in an optimal configur<strong>at</strong>ion for substr<strong>at</strong>e<br />
interaction with the active site <strong>of</strong> Ubc9 <strong>and</strong> for<br />
c<strong>at</strong>alysis. Notably, the IR domain <strong>of</strong> RanBP2<br />
binds extremely stably to Ubc9 <strong>and</strong> the<br />
SUMO-1-conjug<strong>at</strong>ed form <strong>of</strong> RanGAP1,<br />
the activ<strong>at</strong>ing protein for the GTPase Ran<br />
(M<strong>at</strong>unis et al., 1998; Saitoh et al., 1998).<br />
Structural analysis yielded the puzzling result
<strong>Journal</strong> <strong>of</strong> <strong>Cell</strong> Science<br />
th<strong>at</strong> this binding abolishes the ability <strong>of</strong> RanBP2<br />
to promote multiple rounds <strong>of</strong> target<br />
<strong>SUMOyl<strong>at</strong>ion</strong> (Reverter <strong>and</strong> Lima, 2005). It<br />
will clearly be important to establish how this<br />
inhibition is overcome for RanBP2 to act as an<br />
E3 enzyme in its physiological context.<br />
Other SUMO ligases<br />
Additional proteins th<strong>at</strong> have been reported as<br />
potential SUMO ligases include histone<br />
deacetylase 4 (HDAC4), KRAB-associ<strong>at</strong>ed<br />
protein 1 (KPA1), Pc2 <strong>and</strong> Topors.<br />
HDAC4 is a histone deacetylase th<strong>at</strong> is a<br />
<strong>SUMOyl<strong>at</strong>ion</strong> target. HDAC4 expression<br />
enhances the <strong>SUMOyl<strong>at</strong>ion</strong> <strong>of</strong> myocyte-specific<br />
enhancer factor 2 (MEF2), as well as other<br />
targets (Geiss-Friedl<strong>and</strong>er <strong>and</strong> Melchior, 2007;<br />
Zhao et al., 2005). HDAC4 can bind to Ubc9,<br />
suggesting th<strong>at</strong> it acts as an E3 enzyme. It has<br />
altern<strong>at</strong>ively been proposed th<strong>at</strong> HDAC4<br />
enhances <strong>SUMOyl<strong>at</strong>ion</strong> by other means, such<br />
as promoting the phosphoryl<strong>at</strong>ion <strong>of</strong> target<br />
proteins <strong>at</strong> sites adjacent to conjug<strong>at</strong>ed lysine<br />
residues (Yang <strong>and</strong> Gregoire, 2006).<br />
The human co-repressor KRAB-associ<strong>at</strong>ed<br />
protein 1 (KAP1) possesses PHD-finger domains<br />
th<strong>at</strong> c<strong>at</strong>alyze intramolecular <strong>SUMOyl<strong>at</strong>ion</strong> <strong>of</strong> an<br />
adjacent KAP1 bromodomain (Peng <strong>and</strong><br />
Wysocka, 2008). <strong>SUMOyl<strong>at</strong>ion</strong> stabilizes the<br />
associ<strong>at</strong>ion <strong>of</strong> the bromodomain with<br />
the chrom<strong>at</strong>in modifiers, thus promoting the<br />
establishment <strong>of</strong> gene silencing. Structural<br />
analysis suggests th<strong>at</strong> the PHD finger <strong>and</strong> the<br />
bromodomain cooper<strong>at</strong>e as an integr<strong>at</strong>ed unit to<br />
recruit Ubc9 <strong>and</strong> facilit<strong>at</strong>e <strong>SUMOyl<strong>at</strong>ion</strong> (Zeng<br />
et al., 2008).<br />
Mammalian Pc2 is a polycomb-group<br />
protein th<strong>at</strong> can act as a SUMO ligase for the<br />
transcriptional co-repressor CtBP (Wotton <strong>and</strong><br />
Merrill, 2007). Pc2 can bind to both Ubc9 <strong>and</strong> its<br />
conjug<strong>at</strong>ion targets, <strong>and</strong> it seems to have a r<strong>at</strong>her<br />
limited spectrum <strong>of</strong> targets.<br />
Topors is a RING-finger protein th<strong>at</strong> binds<br />
DNA topoisomerase I <strong>and</strong> p53. It possesses both<br />
RING-finger-dependent ubiquitin ligase<br />
activity <strong>and</strong> RING-finger-independent SUMO<br />
ligase activity (Weger et al., 2005). Topors has a<br />
SIM (Hecker et al., 2006) <strong>and</strong> acts as a SUMO<br />
ligase in vitro (Hammer et al., 2007).<br />
SUMO-deconjug<strong>at</strong>ing enzymes<br />
Ulps/SENPs are responsible both for processing<br />
SUMO peptides <strong>and</strong> for deconjug<strong>at</strong>ing<br />
SUMOyl<strong>at</strong>ed species (Hay, 2007;<br />
Mukhopadhyay <strong>and</strong> Dasso, 2007). Ulps/SENPs<br />
share a conserved ~200-amino-acid c<strong>at</strong>alytic<br />
domain th<strong>at</strong> is typically found near their<br />
C-terminus.<br />
There are two Ulps in budding yeast: Ulp1p<br />
<strong>and</strong> Ulp2p (Li <strong>and</strong> Hochstrasser, 1999; Li <strong>and</strong><br />
Hochstrasser, 2000). Ulp1p localizes to the<br />
nuclear envelope <strong>and</strong> is encoded by an<br />
essential gene (Li <strong>and</strong> Hochstrasser, 1999).<br />
Overexpression <strong>of</strong> processed Smt3p weakly<br />
rescues �ulp1 cells, but unprocessed Smt3p does<br />
not (Li <strong>and</strong> Hochstrasser, 1999), suggesting th<strong>at</strong><br />
one essential function <strong>of</strong> Ulp1p is Smt3p<br />
m<strong>at</strong>ur<strong>at</strong>ion. Ulp2p localizes in the nucleoplasm<br />
(Li <strong>and</strong> Hochstrasser, 2000) <strong>and</strong> is particularly<br />
important for dismantling poly-Smt3p chains<br />
(Bylebyl et al., 2003). Although not essential for<br />
veget<strong>at</strong>ive growth, Ulp2p has roles in<br />
chromosome segreg<strong>at</strong>ion, meiotic development<br />
<strong>and</strong> recovery from cell-cycle checkpoint arrest<br />
(Li <strong>and</strong> Hochstrasser, 2000).<br />
Mammals have six SENPs: SENP1, SENP2,<br />
SENP3, SENP5, SENP6 <strong>and</strong> SENP7. SENP1-3<br />
<strong>and</strong> SENP5 are more similar to Ulp1p than to<br />
Ulp2p, whereas SENP6 <strong>and</strong> SENP7 are more<br />
Ulp2-like (Mukhopadhyay <strong>and</strong> Dasso, 2007).<br />
SENP1 <strong>and</strong> SENP2 localize to the nuclear<br />
envelope <strong>and</strong> have processing <strong>and</strong><br />
deconjug<strong>at</strong>ion activity for both SUMO-1 <strong>and</strong><br />
SUMO-2/3. By contrast, all other SENPs have a<br />
strong preference for SUMO-2/3. SENP3 <strong>and</strong><br />
SENP5 localize in nucleoli <strong>and</strong> c<strong>at</strong>alyze<br />
SUMO-2/3 processing <strong>and</strong> deconjug<strong>at</strong>ion (Di<br />
Bacco et al., 2006; Gong <strong>and</strong> Yeh, 2006).<br />
Similar to Ulp1p (Panse et al., 2006), SENP3<br />
<strong>and</strong> SENP5 have important roles in ribosome<br />
biogenesis (Yun et al., 2008). SENP6 <strong>and</strong><br />
SENP7 localize within the nucleoplasm <strong>and</strong> are<br />
implic<strong>at</strong>ed in the editing <strong>of</strong> poly-SUMO chains<br />
(Lima <strong>and</strong> Reverter, 2008; Mukhopadhyay<br />
et al., 2006; Shen et al., 2009).<br />
The sites <strong>of</strong> SENP1, SENP2 <strong>and</strong> Ulp1p th<strong>at</strong> are<br />
engaged during processing or deconjug<strong>at</strong>ion are<br />
shallow clefts lined with conserved amino acids<br />
(Mossessova <strong>and</strong> Lima, 2000; Reverter <strong>and</strong><br />
Lima, 2004; Shen et al., 2006b). In both reactions,<br />
the C-terminus <strong>of</strong> SUMO lies within these clefts<br />
as an elong<strong>at</strong>ed str<strong>and</strong>, <strong>and</strong> conserved<br />
tryptophans <strong>and</strong> other adjacent residues <strong>of</strong> Ulp1p,<br />
SENP1 <strong>and</strong> SENP2 clamp the diglycine motif <strong>of</strong><br />
SUMO in a hydrophobic ‘tunnel’. During<br />
deconjug<strong>at</strong>ion, such binding requires minimal<br />
structural distortion <strong>of</strong> the target protein, which<br />
explains how Ulps/SENPs can deconjug<strong>at</strong>e many<br />
SUMOyl<strong>at</strong>ed species with only modest target<br />
specificity. In the SUMO processing reaction,<br />
Ulps/SENPs induce the isomeriz<strong>at</strong>ion <strong>of</strong> the<br />
scissile peptide bond, resulting in a 90° kink in the<br />
SUMO C-terminal tail (Reverter <strong>and</strong> Lima, 2006;<br />
Shen et al., 2006a). In deconjug<strong>at</strong>ion reactions,<br />
Ulps/SENPs induce the scissile isopeptide bond<br />
between the C-terminus <strong>of</strong> SUMO <strong>and</strong> the<br />
�-amine group <strong>of</strong> the lysine residue <strong>of</strong> the target<br />
protein to adopt a cis configur<strong>at</strong>ion, resulting in a<br />
similar 90° kink (Reverter <strong>and</strong> Lima, 2006; Shen<br />
et al., 2006a). For both peptide <strong>and</strong> amide bonds,<br />
the kinked cis conform<strong>at</strong>ions facilit<strong>at</strong>e hydrolysis<br />
<strong>of</strong> the bond.<br />
<strong>Journal</strong> <strong>of</strong> <strong>Cell</strong> Science 122 (23)<br />
4251<br />
The f<strong>at</strong>es <strong>of</strong> SUMOyl<strong>at</strong>ed species<br />
The consequences <strong>of</strong> <strong>SUMOyl<strong>at</strong>ion</strong> are diverse,<br />
including alter<strong>at</strong>ion <strong>of</strong> the activity, localiz<strong>at</strong>ion<br />
<strong>and</strong>/or stability <strong>of</strong> the target protein (Geiss-<br />
Friedl<strong>and</strong>er <strong>and</strong> Melchior, 2007). Frequently,<br />
these consequences result from recognition <strong>of</strong><br />
conjug<strong>at</strong>ed species by SIM-containing proteins.<br />
<strong>SUMOyl<strong>at</strong>ion</strong> might also cause the loss <strong>of</strong><br />
binding partners, or cause conform<strong>at</strong>ional<br />
changes th<strong>at</strong> alter the enzym<strong>at</strong>ic activity <strong>of</strong> the<br />
target protein. Some SIM-medi<strong>at</strong>ed interactions<br />
prevent deconjug<strong>at</strong>ion by limiting the access <strong>of</strong><br />
Ulps/SENPs to the conjug<strong>at</strong>ed proteins. In cases<br />
in which the interacting SIMs have intrinsic<br />
paralogue preference, they will selectively<br />
protect targets th<strong>at</strong> are modified with the<br />
preferred SUMO paralogue (Zhu et al., 2009).<br />
Few <strong>SUMOyl<strong>at</strong>ion</strong> targets show quantit<strong>at</strong>ive<br />
modific<strong>at</strong>ion. Notably, <strong>SUMOyl<strong>at</strong>ion</strong> can play<br />
an important regul<strong>at</strong>ory role even under these<br />
circumstances (Geiss-Friedl<strong>and</strong>er <strong>and</strong> Melchior,<br />
2007). This might be explained by the<br />
observ<strong>at</strong>ion th<strong>at</strong> <strong>SUMOyl<strong>at</strong>ion</strong> can promote<br />
the assembly <strong>of</strong> protein complexes, such as in<br />
transcriptionally repressed chrom<strong>at</strong>in, th<strong>at</strong><br />
remain stable despite subsequent<br />
de<strong>SUMOyl<strong>at</strong>ion</strong> (Geiss-Friedl<strong>and</strong>er <strong>and</strong><br />
Melchior, 2007). Additionally, <strong>SUMOyl<strong>at</strong>ion</strong><br />
might function within the c<strong>at</strong>alytic cycle <strong>of</strong><br />
targets, facilit<strong>at</strong>ing enzym<strong>at</strong>ic turnover<br />
(Hardel<strong>and</strong> et al., 2002). In both <strong>of</strong> these cases,<br />
SUMOyl<strong>at</strong>ed species are transient intermedi<strong>at</strong>es<br />
th<strong>at</strong> facilit<strong>at</strong>e stable changes in target proteins.<br />
Crosstalk between the SUMO <strong>and</strong><br />
ubiquitin p<strong>at</strong>hways<br />
A particularly exciting development in this field<br />
has revealed an important point <strong>of</strong> crosstalk<br />
between the SUMO <strong>and</strong> ubiquitin p<strong>at</strong>hways<br />
(Hunter <strong>and</strong> Sun, 2008): a subset <strong>of</strong> targets<br />
become conjug<strong>at</strong>ed with multiple SUMOs, <strong>and</strong><br />
can be recognized by SUMO-targeted ubiquitin<br />
ligases (STUbLs), causing the proteasomal<br />
degrad<strong>at</strong>ion <strong>of</strong> these targets. It is currently<br />
believed th<strong>at</strong> STUbLs oper<strong>at</strong>e primarily through<br />
recognition <strong>of</strong> poly-SUMO chains, although it<br />
remains possible th<strong>at</strong> they might recognize some<br />
targets th<strong>at</strong> are mono-SUMOyl<strong>at</strong>ed <strong>at</strong> numerous<br />
sites.<br />
Rfp1p <strong>and</strong> Rfp2p are fission-yeast RINGfinger<br />
proteins th<strong>at</strong> possess N-terminal SIMs<br />
(Sun et al., 2007). They are genetically<br />
redundant <strong>and</strong> there is no visible phenotype for<br />
loss <strong>of</strong> either gene encoding these proteins.<br />
However, cells must possess <strong>at</strong> least one <strong>of</strong> these<br />
proteins for growth <strong>and</strong> genome stability. Both<br />
Rfp1p <strong>and</strong> Rfp2p heterodimerize with the Slx8p<br />
protein, a RING-finger ubiquitin ligase.<br />
Together, they medi<strong>at</strong>e the ubiquityl<strong>at</strong>ion <strong>of</strong><br />
poly-SUMOyl<strong>at</strong>ed targets, resulting in their<br />
proteasomal destruction. The Slx5p-Slx8p
<strong>Journal</strong> <strong>of</strong> <strong>Cell</strong> Science<br />
4252<br />
<strong>Journal</strong> <strong>of</strong> <strong>Cell</strong> Science 122 (23)<br />
heterodimer acts similarly in budding yeast<br />
(Hunter <strong>and</strong> Sun, 2008). The functions <strong>of</strong> Rfp1<br />
<strong>and</strong>/or Rfp2 <strong>and</strong> Slx8 are performed by a single<br />
protein in human cells, RING-finger protein 4<br />
(RNF4), which is the only confirmed<br />
mammalian STUbL (Sun et al., 2007).<br />
Perspectives<br />
Findings during the last 5 or 6 years have<br />
provided a much more sophistic<strong>at</strong>ed<br />
underst<strong>and</strong>ing <strong>of</strong> the <strong>SUMOyl<strong>at</strong>ion</strong> p<strong>at</strong>hway,<br />
revealing some aspects th<strong>at</strong> are unique to this<br />
p<strong>at</strong>hway <strong>and</strong> others th<strong>at</strong> are probably common<br />
to p<strong>at</strong>hways involving all ubiquitin-like<br />
proteins. However, the picture <strong>of</strong> <strong>SUMOyl<strong>at</strong>ion</strong><br />
is not yet complete, <strong>and</strong> we expect th<strong>at</strong> studies <strong>of</strong><br />
the SUMO p<strong>at</strong>hway may yet hold more<br />
surprises. In particular, we look forward to<br />
future findings regarding the mechanisms <strong>of</strong><br />
SUMO ligases <strong>and</strong> the possibility th<strong>at</strong> additional<br />
SIMs remain to be discovered. Finally, we still<br />
have much to learn regarding the biological<br />
functions <strong>of</strong> this p<strong>at</strong>hway <strong>and</strong> its interaction with<br />
ubiquitin <strong>and</strong> other regul<strong>at</strong>ory p<strong>at</strong>hways.<br />
This work was supported through Eunice Kennedy<br />
Shriver N<strong>at</strong>ional Institute <strong>of</strong> Child Health <strong>and</strong> Human<br />
Development Intramural funds (Z01 HD001902).<br />
Deposited in PMC for release after 12 months.<br />
References<br />
Ayaydin, F. <strong>and</strong> Dasso, M. (2004). Distinct in vivo dynamics <strong>of</strong><br />
vertebr<strong>at</strong>e SUMO paralogues. Mol. Biol. <strong>Cell</strong> 15, 5208-5218.<br />
Azuma, Y., Arnaoutov, A., Anan, T. <strong>and</strong> Dasso, M. (2005).<br />
PIASy medi<strong>at</strong>es SUMO-2 conjug<strong>at</strong>ion <strong>of</strong> Topoisomerase-II on<br />
mitotic chromosomes. EMBO J. 24, 2172-2182.<br />
Bernier-Villamor, V., Sampson, D. A., M<strong>at</strong>unis, M. J. <strong>and</strong><br />
Lima, C. D. (2002). Structural basis for E2-medi<strong>at</strong>ed SUMO<br />
conjug<strong>at</strong>ion revealed by a complex between ubiquitinconjug<strong>at</strong>ing<br />
enzyme Ubc9 <strong>and</strong> RanGAP1. <strong>Cell</strong> 108, 345-356.<br />
Bylebyl, G. R., Belichenko, I. <strong>and</strong> Johnson, E. S. (2003). The<br />
SUMO isopeptidase Ulp2 prevents accumul<strong>at</strong>ion <strong>of</strong> SUMO<br />
chains in yeast. J. Biol. Chem. 278, 44113-44120.<br />
Cheng, C. H., Lo, Y. H., Liang, S. S., Ti, S. C., Lin, F. M., Yeh,<br />
C. H., Huang, H. Y. <strong>and</strong> Wang, T. F. (2006). SUMO<br />
modific<strong>at</strong>ions control assembly <strong>of</strong> synaptonemal complex <strong>and</strong><br />
polycomplex in meiosis <strong>of</strong> Saccharomyces cerevisiae. Genes Dev.<br />
20, 2067-2081.<br />
Di Bacco, A., Ouyang, J., Lee, H. Y., C<strong>at</strong>ic, A., Ploegh, H. <strong>and</strong><br />
Gill, G. (2006). The SUMO-specific protease SENP5 is required<br />
for cell division. Mol. <strong>Cell</strong>. Biol. 26, 4489-4498.<br />
Geiss-Friedl<strong>and</strong>er, R. <strong>and</strong> Melchior, F. (2007). Concepts in<br />
sumoyl<strong>at</strong>ion: a decade on. N<strong>at</strong>. Rev Mol. <strong>Cell</strong>. Biol. 8, 947-956.<br />
Gong, L. <strong>and</strong> Yeh, E. T. (2006). Characteriz<strong>at</strong>ion <strong>of</strong> a family <strong>of</strong><br />
nucleolar SUMO-specific proteases with preference for SUMO-<br />
2 or SUMO-3. J. Biol. Chem. 281, 15869-15877.<br />
Grabbe, C. <strong>and</strong> Dikic, I. (2009). Functional roles <strong>of</strong> ubiquitinlike<br />
domain (ULD) <strong>and</strong> ubiquitin-binding domain (UBD)<br />
containing proteins. Chem. Rev. 109, 1481-1494.<br />
Hammer, E., Heilbronn, R. <strong>and</strong> Weger, S. (2007). The E3 ligase<br />
Topors induces the accumul<strong>at</strong>ion <strong>of</strong> polysumoyl<strong>at</strong>ed forms <strong>of</strong><br />
DNA topoisomerase I in vitro <strong>and</strong> in vivo. FEBS Lett. 581, 5418-<br />
5424.<br />
Hardel<strong>and</strong>, U., Steinacher, R., Jiricny, J. <strong>and</strong> Schar, P. (2002).<br />
Modific<strong>at</strong>ion <strong>of</strong> the human thymine-DNA glycosylase by<br />
ubiquitin-like proteins facilit<strong>at</strong>es enzym<strong>at</strong>ic turnover. EMBO J.<br />
21, 1456-1464.<br />
Hay, R. T. (2007). SUMO-specific proteases: a twist in the tail.<br />
Trends <strong>Cell</strong> Biol. 17, 370-376.<br />
Hecker, C. M., Rabiller, M., Haglund, K., Bayer, P. <strong>and</strong> Dikic,<br />
I. (2006). Specific<strong>at</strong>ion <strong>of</strong> SUMO1- <strong>and</strong> SUMO2-interacting<br />
motifs. J. Biol. Chem. 281, 16117-16127.<br />
Hochstrasser, M. (2001). SP-RING for SUMO: new functions<br />
bloom for a ubiquitin-like protein. <strong>Cell</strong> 107, 5-8.<br />
Hochstrasser, M. (2009). Origin <strong>and</strong> function <strong>of</strong> ubiquitin-like<br />
proteins. N<strong>at</strong>ure 458, 422-429.<br />
Hunter, T. <strong>and</strong> Sun, H. (2008). Crosstalk between the SUMO<br />
<strong>and</strong> ubiquitin p<strong>at</strong>hways. Ernst Schering Found. Symp. Proc. 1-16.<br />
Johnson, E. S. (2004). Protein modific<strong>at</strong>ion by SUMO. Annu.<br />
Rev. Biochem. 73, 355-382.<br />
Johnson, E. S. <strong>and</strong> Gupta, A. A. (2001). An E3-like factor th<strong>at</strong><br />
promotes SUMO conjug<strong>at</strong>ion to the yeast septins. <strong>Cell</strong> 106, 735-<br />
744.<br />
Johnson, E. S., Schwienhorst, I., Dohmen, R. J. <strong>and</strong> Blobel,<br />
G. (1997). The ubiquitin-like protein Smt3p is activ<strong>at</strong>ed for<br />
conjug<strong>at</strong>ion to other proteins by an Aos1p/Uba2p heterodimer.<br />
EMBO J. 16, 5509-5519.<br />
Kerscher, O. (2007). SUMO junction-wh<strong>at</strong>’s your function? New<br />
insights through SUMO-interacting motifs. EMBO Rep. 8, 550-<br />
555.<br />
Knipscheer, P., Flotho, A., Klug, H., Olsen, J. V., van Dijk, W.<br />
J., Fish, A., Johnson, E. S., Mann, M., Sixma, T. K. <strong>and</strong><br />
Pichler, A. (2008). Ubc9 sumoyl<strong>at</strong>ion regul<strong>at</strong>es SUMO target<br />
discrimin<strong>at</strong>ion. Mol. <strong>Cell</strong> 31, 371-382.<br />
Li, S. J. <strong>and</strong> Hochstrasser, M. (1999). A new protease required<br />
for cell-cycle progression in yeast. N<strong>at</strong>ure 398, 246-251.<br />
Li, S. J. <strong>and</strong> Hochstrasser, M. (2000). The yeast ULP2 (SMT4)<br />
gene encodes a novel protease specific for the ubiquitin-like Smt3<br />
protein. Mol. <strong>Cell</strong>. Biol. 20, 2367-2377.<br />
Lima, C. D. <strong>and</strong> Reverter, D. (2008). Structure <strong>of</strong> the human<br />
SENP7 c<strong>at</strong>alytic domain <strong>and</strong> poly-SUMO deconjug<strong>at</strong>ion<br />
activities for SENP6 <strong>and</strong> SENP7. J. Biol. Chem. 283, 32045-<br />
32055.<br />
Lois, L. M. <strong>and</strong> Lima, C. D. (2005). Structures <strong>of</strong> the SUMO<br />
E1 provide mechanistic insights into SUMO activ<strong>at</strong>ion <strong>and</strong> E2<br />
recruitment to E1. EMBO J. 24, 439-451.<br />
M<strong>at</strong>ic, I., van Hagen, M., Schimmel, J., Macek, B., Ogg, S. C.,<br />
T<strong>at</strong>ham, M. H., Hay, R. T., Lamond, A. I., Mann, M. <strong>and</strong><br />
Vertegaal, A. C. (2008). In vivo identific<strong>at</strong>ion <strong>of</strong> human small<br />
ubiquitin-like modifier polymeriz<strong>at</strong>ion sites by high accuracy<br />
mass spectrometry <strong>and</strong> an in vitro to in vivo str<strong>at</strong>egy. Mol. <strong>Cell</strong><br />
Proteomics 7, 132-144.<br />
M<strong>at</strong>unis, M. J., Wu, J. <strong>and</strong> Blobel, G. (1998). SUMO-1<br />
modific<strong>at</strong>ion <strong>and</strong> its role in targeting the Ran GTPase-activ<strong>at</strong>ing<br />
protein, RanGAP1, to the nuclear pore complex. J. <strong>Cell</strong> Biol. 140,<br />
499-509.<br />
Meulmeester, E., Kunze, M., Hsiao, H. H., Urlaub, H. <strong>and</strong><br />
Melchior, F. (2008). Mechanism <strong>and</strong> consequences for paralogspecific<br />
sumoyl<strong>at</strong>ion <strong>of</strong> ubiquitin-specific protease 25. Mol. <strong>Cell</strong><br />
30, 610-669.<br />
Mossessova, E. <strong>and</strong> Lima, C. D. (2000). Ulp1-SUMO crystal<br />
structure <strong>and</strong> genetic analysis reveal conserved interactions <strong>and</strong> a<br />
regul<strong>at</strong>ory element essential for cell growth in yeast. Mol. <strong>Cell</strong> 5,<br />
865-876.<br />
Mukhopadhyay, D. <strong>and</strong> Dasso, M. (2007). Modific<strong>at</strong>ion in<br />
reverse: the SUMO proteases. Trends Biochem. Sci. 32, 286-295.<br />
Mukhopadhyay, D., Ayaydin, F., Kolli, N., Tan, S. H., Anan,<br />
T., Kametaka, A., Azuma, Y., Wilkinson, K. D. <strong>and</strong> Dasso, M.<br />
(2006). SUSP1 antagonizes form<strong>at</strong>ion <strong>of</strong> highly SUMO2/3conjug<strong>at</strong>ed<br />
species. J. <strong>Cell</strong> Biol. 174, 939-949.<br />
Ouyang, J., Shi, Y., Valin, A., Xuan, Y. <strong>and</strong> Gill, G. (2009).<br />
Direct binding <strong>of</strong> CoREST1 to SUMO-2/3 contributes to genespecific<br />
repression by the LSD1/CoREST1/HDAC complex.<br />
Mol. <strong>Cell</strong> 34, 145-154.<br />
Palvimo, J. J. (2007). PIAS proteins as regul<strong>at</strong>ors <strong>of</strong> small<br />
ubiquitin-rel<strong>at</strong>ed modifier (SUMO) modific<strong>at</strong>ions <strong>and</strong><br />
transcription. Biochem. Soc. Trans. 35, 1405-1408.<br />
Panse, V. G., Kressler, D., Pauli, A., Petfalski, E., Gnadig, M.,<br />
Tollervey, D. <strong>and</strong> Hurt, E. (2006). Form<strong>at</strong>ion <strong>and</strong> nuclear export<br />
<strong>of</strong> preribosomes are functionally linked to the small-ubiquitinrel<strong>at</strong>ed<br />
modifier p<strong>at</strong>hway. Traffic 7, 1311-1321.<br />
Peng, J. <strong>and</strong> Wysocka, J. (2008). It takes a PHD to SUMO.<br />
Trends Biochem. Sci. 33, 191-194.<br />
Pichler, A., Gast, A., Seeler, J. S., Dejean, A. <strong>and</strong> Melchior, F.<br />
(2002). The nucleoporin RanBP2 has SUMO1 E3 ligase activity.<br />
<strong>Cell</strong> 108, 109-120.<br />
Pickart, C. M. <strong>and</strong> Fushman, D. (2004). Polyubiquitin chains:<br />
polymeric protein signals. Curr. Opin. Chem. Biol. 8, 610-616.<br />
Potts, P. R. (2009). The Yin <strong>and</strong> Yang <strong>of</strong> the MMS21-SMC5/6<br />
SUMO ligase complex in homologous recombin<strong>at</strong>ion. DNA<br />
Repair (Amst) 8, 499-506.<br />
Reverter, D. <strong>and</strong> Lima, C. D. (2004). A basis for SUMO<br />
protease specificity provided by analysis <strong>of</strong> human Senp2 <strong>and</strong> a<br />
Senp2-SUMO complex. Structure 12, 1519-1531.<br />
Reverter, D. <strong>and</strong> Lima, C. D. (2005). Insights into E3 ligase<br />
activity revealed by a SUMO-RanGAP1-Ubc9-Nup358 complex.<br />
N<strong>at</strong>ure 435, 687-692.<br />
Reverter, D. <strong>and</strong> Lima, C. D. (2006). Structural basis for SENP2<br />
protease interactions with SUMO precursors <strong>and</strong> conjug<strong>at</strong>ed<br />
substr<strong>at</strong>es. N<strong>at</strong>. Struct. Mol. Biol. 13, 1060-1068.<br />
Saitoh, H. <strong>and</strong> Hinchey, J. (2000). Functional heterogeneity <strong>of</strong><br />
small ubiquitin-rel<strong>at</strong>ed protein modifiers SUMO-1 versus<br />
SUMO-2/3. J. Biol. Chem. 275, 6252-6258.<br />
Saitoh, H., Sparrow, D. B., Shiomi, T., Pu, R. T., Nishimoto,<br />
T., Mohun, T. J. <strong>and</strong> Dasso, M. (1998). Ubc9p <strong>and</strong> the<br />
conjug<strong>at</strong>ion <strong>of</strong> SUMO-1 to RanGAP1 <strong>and</strong> RanBP2. Curr. Biol.<br />
8, 121-124.<br />
Shen, L., T<strong>at</strong>ham, M. H., Dong, C., Zagorska, A., Naismith,<br />
J. H. <strong>and</strong> Hay, R. T. (2006a). SUMO protease SENP1 induces<br />
isomeriz<strong>at</strong>ion <strong>of</strong> the scissile peptide bond. N<strong>at</strong>. Struct. Mol. Biol.<br />
13, 1069-1077.<br />
Shen, L. N., Dong, C., Liu, H., Naismith, J. H. <strong>and</strong> Hay, R. T.<br />
(2006b). The structure <strong>of</strong> SENP1-SUMO-2 complex suggests a<br />
structural basis for discrimin<strong>at</strong>ion between SUMO paralogues<br />
during processing. Biochem. J. 397, 279-288.<br />
Shen, L. N., Ge<strong>of</strong>froy, M. C., Jaffray, E. G. <strong>and</strong> Hay, R. T.<br />
(2009). Characteriz<strong>at</strong>ion <strong>of</strong> SENP7, a SUMO-2/-3 specific<br />
isopeptidase. Biochem. J. 2, 223-230.<br />
Sun, H., Leverson, J. D. <strong>and</strong> Hunter, T. (2007). Conserved<br />
function <strong>of</strong> RNF4 family proteins in eukaryotes: targeting a<br />
ubiquitin ligase to SUMOyl<strong>at</strong>ed proteins. EMBO J. 26, 4102-<br />
4112.<br />
Takahashi, Y., Kahyo, T., Toh, E. A., Yasuda, H. <strong>and</strong> Kikuchi,<br />
Y. (2001). Yeast Ull1/Siz1 is a novel SUMO1/Smt3 ligase for<br />
septin components <strong>and</strong> functions as an adaptor between<br />
conjug<strong>at</strong>ing enzyme <strong>and</strong> substr<strong>at</strong>es. J. Biol. Chem. 276, 48973-<br />
48977.<br />
T<strong>at</strong>ham, M. H., Jaffray, E., Vaughan, O. A., Desterro, J. M.,<br />
Botting, C. H., Naismith, J. H. <strong>and</strong> Hay, R. T. (2001).<br />
Polymeric chains <strong>of</strong> SUMO-2 <strong>and</strong> SUMO-3 are conjug<strong>at</strong>ed to<br />
protein substr<strong>at</strong>es by SAE1/SAE2 <strong>and</strong> Ubc9. J. Biol. Chem. 276,<br />
35368-35374.<br />
Vertegaal, A. C., Andersen, J. S., Ogg, S. C., Hay, R. T., Mann,<br />
M. <strong>and</strong> Lamond, A. I. (2006). Distinct <strong>and</strong> overlapping sets <strong>of</strong><br />
SUMO-1 <strong>and</strong> SUMO-2 target proteins revealed by quantit<strong>at</strong>ive<br />
proteomics. Mol. <strong>Cell</strong> Proteomics 5, 2298-2310.<br />
Weger, S., Hammer, E. <strong>and</strong> Heilbronn, R. (2005). Topors acts<br />
as a SUMO-1 E3 ligase for p53 in vitro <strong>and</strong> in vivo. FEBS Lett.<br />
579, 5007-5012.<br />
Wotton, D. <strong>and</strong> Merrill, J. C. (2007). Pc2 <strong>and</strong> <strong>SUMOyl<strong>at</strong>ion</strong>.<br />
Biochem. Soc. Trans. 35, 1401-1404.<br />
Yang, X. J. <strong>and</strong> Gregoire, S. (2006). A recurrent phosphosumoyl<br />
switch in transcriptional repression <strong>and</strong> beyond. Mol. <strong>Cell</strong><br />
23, 779-786.<br />
Yun, C., Wang, Y., Mukhopadhyay, D., Backlund, P., Kolli,<br />
N., Yergey, A., Wilkinson, K. D. <strong>and</strong> Dasso, M. (2008).<br />
Nucleolar protein B23/nucleophosmin regul<strong>at</strong>es the vertebr<strong>at</strong>e<br />
SUMO p<strong>at</strong>hway through SENP3 <strong>and</strong> SENP5 proteases. J. <strong>Cell</strong><br />
Biol. 183, 589-595.<br />
Yunus, A. A. <strong>and</strong> Lima, C. D. (2006). Lysine activ<strong>at</strong>ion <strong>and</strong><br />
functional analysis <strong>of</strong> E2-medi<strong>at</strong>ed conjug<strong>at</strong>ion in the SUMO<br />
p<strong>at</strong>hway. N<strong>at</strong>. Struct. Mol. Biol. 13, 491-499.<br />
Yunus, A. A. <strong>and</strong> Lima, C. D. (2009). Structure <strong>of</strong> the Siz/PIAS<br />
SUMO E3 ligase Siz1 <strong>and</strong> determinants required for SUMO<br />
modific<strong>at</strong>ion <strong>of</strong> PCNA. Mol. <strong>Cell</strong> 35, 669-682.<br />
Zeng, L., Yap, K. L., Ivanov, A. V., Wang, X., Mujtaba, S.,<br />
Plotnikova, O., Rauscher, F. J., 3rd. <strong>and</strong> Zhou, M. M.<br />
(2008). Structural insights into human KAP1 PHD fingerbromodomain<br />
<strong>and</strong> its role in gene silencing. N<strong>at</strong>. Struct. Mol. Biol.<br />
15, 626-633.<br />
Zhang, X. D., Goeres, J., Zhang, H., Yen, T. J., Porter, A. C.<br />
<strong>and</strong> M<strong>at</strong>unis, M. J. (2008). SUMO-2/3 modific<strong>at</strong>ion <strong>and</strong> binding<br />
regul<strong>at</strong>e the associ<strong>at</strong>ion <strong>of</strong> CENP-E with kinetochores <strong>and</strong><br />
progression through mitosis. Mol. <strong>Cell</strong> 29, 729-741.<br />
Zhao, X., Sternsdorf, T., Bolger, T. A., Evans, R. M. <strong>and</strong> Yao,<br />
T. P. (2005). Regul<strong>at</strong>ion <strong>of</strong> MEF2 by histone deacetylase 4- <strong>and</strong><br />
SIRT1 deacetylase-medi<strong>at</strong>ed lysine modific<strong>at</strong>ions. Mol. <strong>Cell</strong>.<br />
Biol. 25, 8456-8464.<br />
Zhu, J., Zhu, S., Guzzo, C. M., Ellis, N. A., Sung, K. S., Choi,<br />
C. Y. <strong>and</strong> M<strong>at</strong>unis, M. J. (2008). Small ubiquitin-rel<strong>at</strong>ed<br />
modifier (SUMO) binding determines substr<strong>at</strong>e recognition <strong>and</strong><br />
paralog-selective SUMO modific<strong>at</strong>ion. J. Biol. Chem. 283,<br />
29405-29415.<br />
Zhu, S., Goeres, J., Sixt, K. M., Bekes, M., Zhang, X. D.,<br />
Salvesen, G. S. <strong>and</strong> M<strong>at</strong>unis, M. J. (2009). Protection from<br />
isopeptidase-medi<strong>at</strong>ed deconjug<strong>at</strong>ion regul<strong>at</strong>es paralog-selective<br />
sumoyl<strong>at</strong>ion <strong>of</strong> RanGAP1. Mol. <strong>Cell</strong> 33, 570-580.<br />
<strong>Cell</strong> Science <strong>at</strong> a <strong>Glance</strong> on the Web<br />
Electronic copies <strong>of</strong> the poster insert are available<br />
in the online version <strong>of</strong> this article <strong>at</strong><br />
jcs.biologists.org. The JPEG images can be<br />
downloaded for printing or used as slides.