Bioinformatics Biocomputing - Ercim
Bioinformatics Biocomputing - Ercim
Bioinformatics Biocomputing - Ercim
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
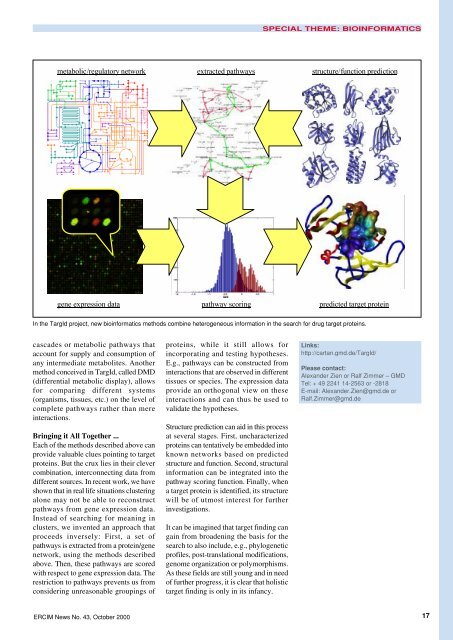
metabolic/regulatory network<br />
gene expression data<br />
cascades or metabolic pathways that<br />
account for supply and consumption of<br />
any intermediate metabolites. Another<br />
method conceived in TargId, called DMD<br />
(differential metabolic display), allows<br />
for comparing different systems<br />
(organisms, tissues, etc.) on the level of<br />
complete pathways rather than mere<br />
interactions.<br />
Bringing it All Together ...<br />
Each of the methods described above can<br />
provide valuable clues pointing to target<br />
proteins. But the crux lies in their clever<br />
combination, interconnecting data from<br />
different sources. In recent work, we have<br />
shown that in real life situations clustering<br />
alone may not be able to reconstruct<br />
pathways from gene expression data.<br />
Instead of searching for meaning in<br />
clusters, we invented an approach that<br />
proceeds inversely: First, a set of<br />
pathways is extracted from a protein/gene<br />
network, using the methods described<br />
above. Then, these pathways are scored<br />
with respect to gene expression data. The<br />
restriction to pathways prevents us from<br />
considering unreasonable groupings of<br />
ERCIM News No. 43, October 2000<br />
extracted pathways<br />
pathway scoring<br />
proteins, while it still allows for<br />
incorporating and testing hypotheses.<br />
E.g., pathways can be constructed from<br />
interactions that are observed in different<br />
tissues or species. The expression data<br />
provide an orthogonal view on these<br />
interactions and can thus be used to<br />
validate the hypotheses.<br />
Structure prediction can aid in this process<br />
at several stages. First, uncharacterized<br />
proteins can tentatively be embedded into<br />
known networks based on predicted<br />
structure and function. Second, structural<br />
information can be integrated into the<br />
pathway scoring function. Finally, when<br />
a target protein is identified, its structure<br />
will be of utmost interest for further<br />
investigations.<br />
It can be imagined that target finding can<br />
gain from broadening the basis for the<br />
search to also include, e.g., phylogenetic<br />
profiles, post-translational modifications,<br />
genome organization or polymorphisms.<br />
As these fields are still young and in need<br />
of further progress, it is clear that holistic<br />
target finding is only in its infancy.<br />
SPECIAL THEME: BIOINFORMATICS<br />
structure/function prediction<br />
predicted target protein<br />
In the TargId project, new bioinformatics methods combine heterogeneous information in the search for drug target proteins.<br />
Links:<br />
http://cartan.gmd.de/TargId/<br />
Please contact:<br />
Alexander Zien or Ralf Zimmer – GMD<br />
Tel: + 49 2241 14-2563 or -2818<br />
E-mail: Alexander.Zien@gmd.de or<br />
Ralf.Zimmer@gmd.de<br />
17