Bioinformatics Biocomputing - Ercim
Bioinformatics Biocomputing - Ercim
Bioinformatics Biocomputing - Ercim
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
SPECIAL THEME: BIOINFORMATICS<br />
Co-operative Environments for Genomes Annotation:<br />
from Imagene to Geno-Annot<br />
by Claudine Médigue, Yves Vandenbrouke, François Rechenmann and Alain Viari<br />
‘Imagene’ is a a co-operative computer environment<br />
for the annotation and analysis of genomic sequences<br />
developed in collaboration between INRIA, Université<br />
Paris 6, Institut Pasteur and the ILOG company. The<br />
first version of this software was dedicated to bacterial<br />
In the context of large-scale genomic<br />
sequencing projects the need is growing<br />
for integration of specific sequence<br />
analysis tools within data management<br />
systems. With this aim in view, we have<br />
developed the Imagene co-operative<br />
computer environment dedicated to<br />
automatic sequence annotation and<br />
analysis (http://abraxa.snv.jussieu.fr/<br />
imagene). In this system, biological<br />
knowledge produced in the course of a<br />
genome sequencing project (putative<br />
genes, regulatory signals, etc) together<br />
with the methodological knowledge,<br />
represented by an extensible set of<br />
sequence analysis methods, are uniformly<br />
represented in an object oriented model.<br />
Imagene is the result of a five years<br />
collaboration between INRIA, Université<br />
Paris 6, the Institut Pasteur and the ILOG<br />
company. The system has been<br />
implemented by using an object oriented<br />
model and a co-operative solving engine<br />
provided by ILOG. In Imagene, a global<br />
problem (task) is solved by successive<br />
decompositions into smaller sub-tasks.<br />
During the execution, the various subtasks<br />
are graphically displayed to the user.<br />
In that sense, Imagene is more transparent<br />
to the user than a traditional menu-driven<br />
package for sequence analysis since all<br />
the steps in the resolution are clearly<br />
identified. Moreover, once a task has been<br />
solved, the user can restart it at any point;<br />
the system then keeps track of the<br />
different versions of the execution. This<br />
allows to maintain several hypothesis in<br />
parallel during the analysis. Imagene also<br />
provides a user interface to display, on<br />
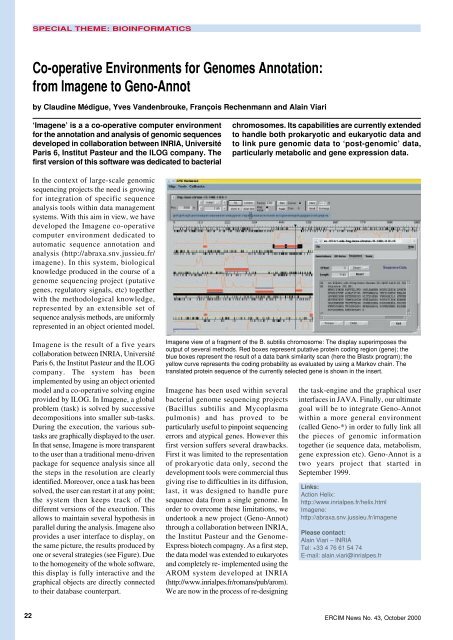
the same picture, the results produced by<br />
one or several strategies (see Figure). Due<br />
to the homogeneity of the whole software,<br />
this display is fully interactive and the<br />
graphical objects are directly connected<br />
to their database counterpart.<br />
Imagene has been used within several<br />
bacterial genome sequencing projects<br />
(Bacillus subtilis and Mycoplasma<br />
pulmonis) and has proved to be<br />
particularly useful to pinpoint sequencing<br />
errors and atypical genes. However this<br />
first version suffers several drawbacks.<br />
First it was limited to the representation<br />
of prokaryotic data only, second the<br />
development tools were commercial thus<br />
giving rise to difficulties in its diffusion,<br />
last, it was designed to handle pure<br />
sequence data from a single genome. In<br />
order to overcome these limitations, we<br />
undertook a new project (Geno-Annot)<br />
through a collaboration between INRIA,<br />
the Institut Pasteur and the Genome-<br />
Express biotech compagny. As a first step,<br />
the data model was extended to eukaryotes<br />
and completely re- implemented using the<br />
AROM system developed at INRIA<br />
(http://www.inrialpes.fr/romans/pub/arom).<br />
We are now in the process of re-designing<br />
chromosomes. Its capabilities are currently extended<br />
to handle both prokaryotic and eukaryotic data and<br />
to link pure genomic data to ‘post-genomic’ data,<br />
particularly metabolic and gene expression data.<br />
Imagene view of a fragment of the B. subtilis chromosome: The display superimposes the<br />
output of several methods. Red boxes represent putative protein coding region (gene); the<br />
blue boxes represent the result of a data bank similarity scan (here the Blastx program); the<br />
yellow curve represents the coding probability as evaluated by using a Markov chain. The<br />
translated protein sequence of the currently selected gene is shown in the insert.<br />
the task-engine and the graphical user<br />
interfaces in JAVA. Finally, our ultimate<br />
goal will be to integrate Geno-Annot<br />
within a more general environment<br />
(called Geno-*) in order to fully link all<br />
the pieces of genomic information<br />
together (ie sequence data, metabolism,<br />
gene expression etc). Geno-Annot is a<br />
two years project that started in<br />
September 1999.<br />
Links:<br />
Action Helix:<br />
http://www.inrialpes.fr/helix.html<br />
Imagene:<br />
http://abraxa.snv.jussieu.fr/imagene<br />
Please contact:<br />
Alain Viari – INRIA<br />
Tel: +33 4 76 61 54 74<br />
E-mail: alain.viari@inrialpes.fr<br />
22 ERCIM News No. 43, October 2000