- Page 1: Making the most of phylogeny: Uniqu
- Page 4 and 5: Eingereicht am: . . . . . . . . . .
- Page 7 and 8: Contents Acronyms xi I. Introductio
- Page 9: List of Figures Contents List of Fi
- Page 12 and 13: Acronyms Acronyms xii ITS2 internal
- Page 15 and 16: Part I. Introduction Thesis outline
- Page 17 and 18: Part I. Introduction The phylum Tar
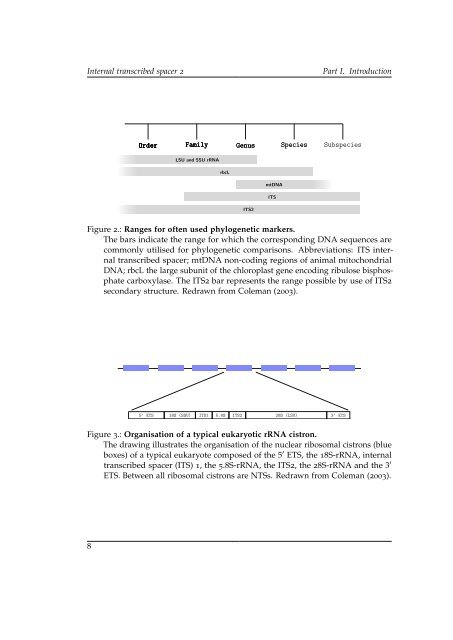
- Page 19: Part I. Introduction Internal trans
- Page 23: Part II. Material and Methods 11
- Page 27 and 28: Chapter 2. Methods 2.1. Bioinformat
- Page 29 and 30: Part II. Material and Methods Bioin
- Page 31 and 32: Part II. Material and Methods Bioin
- Page 33: Part III. Results 21
- Page 36 and 37: BMC Genomics BioMed Central Researc
- Page 38 and 39: BMC Genomics 2009, 10:469 http://ww
- Page 40 and 41: BMC Genomics 2009, 10:469 http://ww
- Page 42 and 43: BMC Genomics 2009, 10:469 http://ww
- Page 44 and 45: BMC Genomics 2009, 10:469 http://ww
- Page 47 and 48: Chapter 4. Transcriptome survey of
- Page 49 and 50: Mali et al. BMC Genomics 2010, 11:1
- Page 51 and 52: Mali et al. BMC Genomics 2010, 11:1
- Page 53 and 54: Mali et al. BMC Genomics 2010, 11:1
- Page 55 and 56: Mali et al. BMC Genomics 2010, 11:1
- Page 57 and 58: Mali et al. BMC Genomics 2010, 11:1
- Page 59 and 60: Chapter 5. Transcriptome analyzed a
- Page 61 and 62: Abstract Tardigrades are unique met
- Page 63 and 64: adiation and cold 17; 18; 19 and H.
- Page 65 and 66: non-directional cDNA library. For t
- Page 67 and 68: available ESTs from both species co
- Page 69 and 70: Individual COG/KOGs: Among the COGs
- Page 71 and 72:
anches, but have only Lea 4 protein
- Page 73 and 74:
emains a question mark in Fig. 5. H
- Page 75 and 76:
(posttranslational modification) an
- Page 77 and 78:
other aspects of the adaptation pro
- Page 79 and 80:
tolerance in man. It is a recombina
- Page 81 and 82:
We used primer3 90 for the design o
- Page 83 and 84:
cathode buffer (25 mM Tris-HCl pH 9
- Page 85 and 86:
24. Ostareck-Lederer, A., Ostareck,
- Page 87 and 88:
59. Gaudermann, P., Vogl, I., Zient
- Page 89 and 90:
91. Bork, P. & Gibson, T. J. (1996)
- Page 91 and 92:
Fig. 2. PCR validation of heat shoc
- Page 93 and 94:
Fig. 7. Comparison of COGs/KOGs spe
- Page 95 and 96:
Table IIa. Subset of important COGs
- Page 97 and 98:
KOG0660:[T]Mitogen-activated protei
- Page 99 and 100:
Table III. Enzymes in metabolic pat
- Page 101 and 102:
ec:5.3.3.2 isopentenyl-diphosphate
- Page 103 and 104:
Table IVa. Specific stress pathway
- Page 111 and 112:
Chapter 6. Proteomic analysis of ta
- Page 113 and 114:
Figure 1. SEM images of M. tardigra
- Page 115 and 116:
listed in Table 2. The individual i
- Page 117 and 118:
Table 1. Overview of identified pro
- Page 119 and 120:
Table 2. Cont. Sequence coverage No
- Page 121 and 122:
Table 2. Cont. Sequence coverage No
- Page 123 and 124:
Table 2. Cont. Sequence coverage No
- Page 125 and 126:
Table 2. Cont. Sequence coverage No
- Page 127 and 128:
Table 2. Cont. Sequence coverage No
- Page 129 and 130:
Table 2. Cont. Sequence coverage No
- Page 131 and 132:
Table 2. Cont. Sequence coverage No
- Page 133 and 134:
Table 2. Cont. Sequence coverage No
- Page 135 and 136:
Table 2. Cont. Sequence coverage No
- Page 137 and 138:
Table 3. Identified proteins withou
- Page 139 and 140:
Table 3. Cont. Spot no. Accession n
- Page 141 and 142:
Table 3. Cont. Spot no. Accession n
- Page 143 and 144:
Figure 8. Detection of hsp60 and hs
- Page 145 and 146:
Unique proteins, when analyzed on t
- Page 147 and 148:
(version 56.1, September 2008, The
- Page 149 and 150:
Chapter 7. Stress response in tardi
- Page 151 and 152:
Due to the remarkable ability of ta
- Page 153 and 154:
Fig. 1 Alignment of two α-crystall
- Page 155 and 156:
active state. This leads to the ass
- Page 157:
Pfaffl MW, Horgan GW, Dempfle L (20
- Page 160 and 161:
Tardigrade bioinformatics: Molecula
- Page 162 and 163:
searched against the next more exte
- Page 164 and 165:
Concerted changes in tardigrade ada
- Page 166 and 167:
mit dynamischen Modellierungstechni
- Page 168 and 169:
Tables and Figures Table 1: Bioinfo
- Page 170 and 171:
Figure 1: Maximum likelihood tree f
- Page 173 and 174:
Chapter 9. The ITS2 Database III—
- Page 175 and 176:
D276 Nucleic Acids Research, 2010,
- Page 177 and 178:
D278 Nucleic Acids Research, 2010,
- Page 179 and 180:
Chapter 10. Including RNA Secondary
- Page 181 and 182:
Keller et al. Biology Direct 2010,
- Page 183 and 184:
Keller et al. Biology Direct 2010,
- Page 185 and 186:
Keller et al. Biology Direct 2010,
- Page 187 and 188:
Keller et al. Biology Direct 2010,
- Page 189 and 190:
Keller et al. Biology Direct 2010,
- Page 191:
Keller et al. Biology Direct 2010,
- Page 194 and 195:
BMC Evolutionary Biology Research a
- Page 196 and 197:
BMC Evolutionary Biology 2008, 8:21
- Page 198 and 199:
BMC Evolutionary Biology 2008, 8:21
- Page 200 and 201:
BMC Evolutionary Biology 2008, 8:21
- Page 202 and 203:
BMC Evolutionary Biology 2008, 8:21
- Page 204 and 205:
BMC Evolutionary Biology 2008, 8:21
- Page 207 and 208:
Chapter 12. Distinguishing species
- Page 209 and 210:
Abstract Distinguishing species in
- Page 211 and 212:
Distinguishing species in Paramacro
- Page 213 and 214:
ITS 2 gene amplification and sequen
- Page 215 and 216:
Distinguishing species in Paramacro
- Page 217 and 218:
Distinguishing species in Paramacro
- Page 219 and 220:
Distinguishing species in Paramacro
- Page 221 and 222:
Distinguishing species in Paramacro
- Page 223 and 224:
References Distinguishing species i
- Page 225 and 226:
Distinguishing species in Paramacro
- Page 227 and 228:
Distinguishing species in Paramacro
- Page 229 and 230:
Distinguishing species in Paramacro
- Page 231 and 232:
Figure legends Distinguishing speci
- Page 233 and 234:
221
- Page 235:
Part IV. General Discussion and Con
- Page 238 and 239:
Part IV. General Discussion and Con
- Page 240 and 241:
Part IV. General Discussion and Con
- Page 242 and 243:
Part IV. General Discussion and Con
- Page 244 and 245:
Part IV. General Discussion and Con
- Page 246 and 247:
Summary succeeded to show the benef
- Page 248 and 249:
Zusammenfassung der internal transc
- Page 250 and 251:
Bibliography Bibliography Bavan, S.
- Page 252 and 253:
Bibliography Bibliography 146.4 (Ap
- Page 254 and 255:
Bibliography Bibliography Horikawa,
- Page 256 and 257:
Bibliography Bibliography Letunic,
- Page 258 and 259:
Bibliography Bibliography Pertea, G
- Page 260 and 261:
Bibliography Bibliography Schultz,
- Page 262 and 263:
Bibliography Bibliography Westh, P.
- Page 264 and 265:
Contributions Markus Grohme wrote t
- Page 266 and 267:
Contributions model and the ITS2 se
- Page 269 and 270:
List of Publications Publications a
- Page 271:
List of Publications Förster, F.,