Checklist of Baltic Sea Macro-species - IOW
Checklist of Baltic Sea Macro-species - IOW
Checklist of Baltic Sea Macro-species - IOW
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
2. <strong>Checklist</strong> documentation and distribution<br />
data for <strong>Baltic</strong> <strong>Sea</strong> macrophyte <strong>species</strong><br />
The checklist for <strong>Baltic</strong> <strong>Sea</strong> macrophyte <strong>species</strong><br />
compiled within the HELCOM Red List project currently<br />
includes 531 taxa <strong>of</strong> macroalgae, aquatic<br />
vascular plants, charophytes and bryophytes.<br />
The list also contains 3052 <strong>of</strong> the most common<br />
synonyms, variations or sub<strong>species</strong>. These data<br />
make it possible to establish the distribution <strong>of</strong><br />
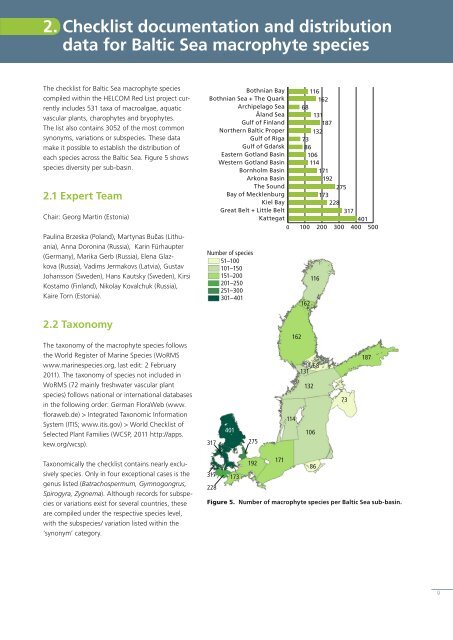
each <strong>species</strong> across the <strong>Baltic</strong> <strong>Sea</strong>. Figure 5 shows<br />
<strong>species</strong> diversity per sub-basin.<br />
2.1 Expert Team<br />
Chair: Georg Martin (Estonia)<br />
Paulina Brzeska (Poland), Martynas Bučas (Lithuania),<br />
Anna Doronina (Russia), Karin Fürhaupter<br />
(Germany), Marika Gerb (Russia), Elena Glazkova<br />
(Russia), Vadims Jermakovs (Latvia), Gustav<br />
Johansson (Sweden), Hans Kautsky (Sweden), Kirsi<br />
Kostamo (Finland), Nikolay Kovalchuk (Russia),<br />
Kaire Torn (Estonia).<br />
Bothnian Bay<br />
Bothnian <strong>Sea</strong> + The Quark<br />
116<br />
162<br />
Archipelago <strong>Sea</strong> 68<br />
Åland <strong>Sea</strong><br />
Gulf <strong>of</strong> Finland<br />
Northern <strong>Baltic</strong> Proper<br />
131<br />
187<br />
132<br />
Gulf <strong>of</strong> Riga<br />
Gulf <strong>of</strong> Gdask<br />
Eastern Gotland Basin<br />
Western Gotland Basin<br />
Bornholm Basin<br />
Arkona Basin<br />
73<br />
86<br />
106<br />
114<br />
171<br />
192<br />
The Sound<br />
275<br />
Bay <strong>of</strong> Mecklenburg<br />
173<br />
Kiel Bay<br />
228<br />
Great Belt + Little Belt<br />
317<br />
Kattegat<br />
401<br />
0 100 200 300 400 500<br />
Number <strong>of</strong> <strong>species</strong><br />
51–100<br />
101–150<br />
151–200<br />
201–250<br />
251–300<br />
301–401<br />
162<br />
116<br />
2.2 Taxonomy<br />
The taxonomy <strong>of</strong> the macrophyte <strong>species</strong> follows<br />
the World Register <strong>of</strong> Marine Species (WoRMS<br />
www.marine<strong>species</strong>.org, last edit: 2 February<br />
2011). The taxonomy <strong>of</strong> <strong>species</strong> not included in<br />
WoRMS (72 mainly freshwater vascular plant<br />
<strong>species</strong>) follows national or international databases<br />
in the following order: German FloraWeb (www.<br />
floraweb.de) > Integrated Taxonomic Information<br />
System (ITIS; www.itis.gov) > World <strong>Checklist</strong> <strong>of</strong><br />
Selected Plant Families (WCSP, 2011 http://apps.<br />
kew.org/wcsp).<br />
317<br />
401<br />
275<br />
162<br />
131 68<br />
132<br />
114<br />
106<br />
73<br />
187<br />
Taxonomically the checklist contains nearly exclusively<br />
<strong>species</strong>. Only in four exceptional cases is the<br />
genus listed (Batrachospermum, Gymnogongrus,<br />
Spirogyra, Zygnema). Although records for sub<strong>species</strong><br />
or variations exist for several countries, these<br />
are compiled under the respective <strong>species</strong> level,<br />
with the sub<strong>species</strong>/ variation listed within the<br />
‘synonym’ category.<br />
171<br />
192<br />
86<br />
317 173<br />
228<br />
Figure 5. Number <strong>of</strong> macrophyte <strong>species</strong> per <strong>Baltic</strong> <strong>Sea</strong> sub-basin.<br />
9