Rapid evolutionary divergence of Photosystem I core subunits PsaA ...
Rapid evolutionary divergence of Photosystem I core subunits PsaA ...
Rapid evolutionary divergence of Photosystem I core subunits PsaA ...
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
136<br />
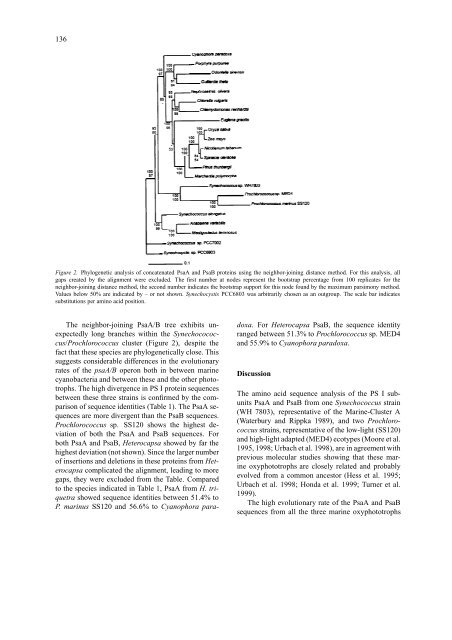
Figure 2. Phylogenetic analysis <strong>of</strong> concatenated <strong>PsaA</strong> and PsaB proteins using the neighbor-joining distance method. For this analysis, all<br />
gaps created by the alignment were excluded. The first number at nodes represent the bootstrap percentage from 100 replicates for the<br />
neighbor-joining distance method, the second number indicates the bootstrap support for this node found by the maximum parsimony method.<br />
Values below 50% are indicated by – or not shown. Synechocystis PCC6803 was arbitrarily chosen as an outgroup. The scale bar indicates<br />
substitutions per amino acid position.<br />
The neighbor-joining <strong>PsaA</strong>/B tree exhibits unexpectedly<br />
long branches within the Synechocococcus/Prochlorococcus<br />
cluster (Figure 2), despite the<br />
fact that these species are phylogenetically close. This<br />
suggests considerable differences in the <strong>evolutionary</strong><br />
rates <strong>of</strong> the psaA/B operon both in between marine<br />
cyanobacteria and between these and the other phototrophs.<br />
The high <strong>divergence</strong> in PS I protein sequences<br />
between these three strains is confirmed by the comparison<br />
<strong>of</strong> sequence identities (Table 1). The <strong>PsaA</strong> sequences<br />
are more divergent than the PsaB sequences.<br />
Prochlorococcus sp. SS120 shows the highest deviation<br />
<strong>of</strong> both the <strong>PsaA</strong> and PsaB sequences. For<br />
both <strong>PsaA</strong> and PsaB, Heterocapsa showed by far the<br />
highest deviation (not shown). Since the larger number<br />
<strong>of</strong> insertions and deletions in these proteins from Heterocapsa<br />
complicated the alignment, leading to more<br />
gaps, they were excluded from the Table. Compared<br />
to the species indicated in Table 1, <strong>PsaA</strong> from H. triquetra<br />
showed sequence identities between 51.4% to<br />
P. marinus SS120 and 56.6% to Cyanophora paradoxa.<br />
For Heterocapsa PsaB, the sequence identity<br />
ranged between 51.3% to Prochlorococcus sp. MED4<br />
and 55.9% to Cyanophora paradoxa.<br />
Discussion<br />
The amino acid sequence analysis <strong>of</strong> the PS I <strong>subunits</strong><br />
<strong>PsaA</strong> and PsaB from one Synechococcus strain<br />
(WH 7803), representative <strong>of</strong> the Marine-Cluster A<br />
(Waterbury and Rippka 1989), and two Prochlorococcus<br />
strains, representative <strong>of</strong> the low-light (SS120)<br />
and high-light adapted (MED4) ecotypes (Moore et al.<br />
1995, 1998; Urbach et al. 1998), are in agreement with<br />
previous molecular studies showing that these marine<br />
oxyphototrophs are closely related and probably<br />
evolved from a common ancestor (Hess et al. 1995;<br />
Urbach et al. 1998; Honda et al. 1999; Turner et al.<br />
1999).<br />
The high <strong>evolutionary</strong> rate <strong>of</strong> the <strong>PsaA</strong> and PsaB<br />
sequences from all the three marine oxyphototrophs