Sumoylation in Aspergillus nidulans: sumO inactivation ...
Sumoylation in Aspergillus nidulans: sumO inactivation ...
Sumoylation in Aspergillus nidulans: sumO inactivation ...
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
K.H. Wong et al. / Fungal Genetics and Biology 45 (2008) 728–737 733<br />
experiments us<strong>in</strong>g the pyroA gene on pI4 (Osmani et al.,<br />
1999) as the selectable marker (data not shown).<br />
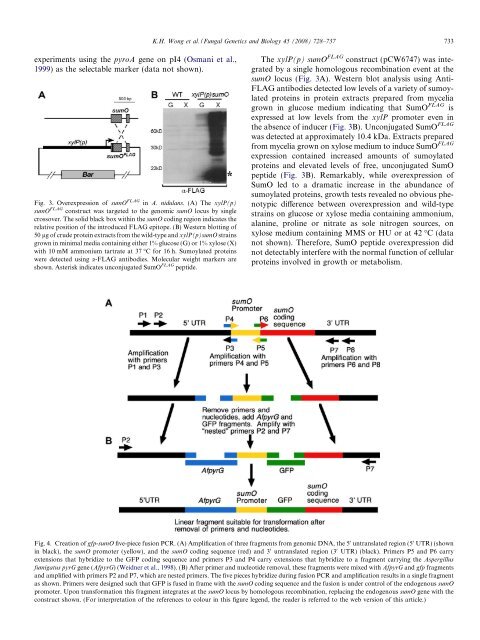
Fig. 3. Overexpression of <strong>sumO</strong> FLAG <strong>in</strong> A. <strong>nidulans</strong>. (A) The xylP(p)<br />
<strong>sumO</strong> FLAG construct was targeted to the genomic <strong>sumO</strong> locus by s<strong>in</strong>gle<br />
crossover. The solid black box with<strong>in</strong> the <strong>sumO</strong> cod<strong>in</strong>g region <strong>in</strong>dicates the<br />
relative position of the <strong>in</strong>troduced FLAG epitope. (B) Western blott<strong>in</strong>g of<br />
50 lg of crude prote<strong>in</strong> extracts from the wild-type and xylP(p)<strong>sumO</strong> stra<strong>in</strong>s<br />
grown <strong>in</strong> m<strong>in</strong>imal media conta<strong>in</strong><strong>in</strong>g either 1% glucose (G) or 1% xylose (X)<br />
with 10 mM ammonium tartrate at 37 °C for 16 h. Sumoylated prote<strong>in</strong>s<br />
were detected us<strong>in</strong>g a-FLAG antibodies. Molecular weight markers are<br />
shown. Asterisk <strong>in</strong>dicates unconjugated SumO FLAG peptide.<br />
The xylP(p) <strong>sumO</strong> FLAG construct (pCW6747) was <strong>in</strong>tegrated<br />
by a s<strong>in</strong>gle homologous recomb<strong>in</strong>ation event at the<br />
<strong>sumO</strong> locus (Fig. 3A). Western blot analysis us<strong>in</strong>g Anti-<br />
FLAG antibodies detected low levels of a variety of sumoylated<br />
prote<strong>in</strong>s <strong>in</strong> prote<strong>in</strong> extracts prepared from mycelia<br />
grown <strong>in</strong> glucose medium <strong>in</strong>dicat<strong>in</strong>g that SumO FLAG is<br />
expressed at low levels from the xylP promoter even <strong>in</strong><br />
the absence of <strong>in</strong>ducer (Fig. 3B). Unconjugated SumO FLAG<br />
was detected at approximately 10.4 kDa. Extracts prepared<br />
from mycelia grown on xylose medium to <strong>in</strong>duce SumO FLAG<br />
expression conta<strong>in</strong>ed <strong>in</strong>creased amounts of sumoylated<br />
prote<strong>in</strong>s and elevated levels of free, unconjugated SumO<br />
peptide (Fig. 3B). Remarkably, while overexpression of<br />
SumO led to a dramatic <strong>in</strong>crease <strong>in</strong> the abundance of<br />
sumoylated prote<strong>in</strong>s, growth tests revealed no obvious phenotypic<br />
difference between overexpression and wild-type<br />
stra<strong>in</strong>s on glucose or xylose media conta<strong>in</strong><strong>in</strong>g ammonium,<br />
alan<strong>in</strong>e, prol<strong>in</strong>e or nitrate as sole nitrogen sources, on<br />
xylose medium conta<strong>in</strong><strong>in</strong>g MMS or HU or at 42 °C (data<br />
not shown). Therefore, SumO peptide overexpression did<br />
not detectably <strong>in</strong>terfere with the normal function of cellular<br />
prote<strong>in</strong>s <strong>in</strong>volved <strong>in</strong> growth or metabolism.<br />
Fig. 4. Creation of gfp-<strong>sumO</strong> five-piece fusion PCR. (A) Amplification of three fragments from genomic DNA, the 5 0 untranslated region (5 0 UTR) (shown<br />
<strong>in</strong> black), the <strong>sumO</strong> promoter (yellow), and the <strong>sumO</strong> cod<strong>in</strong>g sequence (red) and 3 0 untranslated region (3 0 UTR) (black). Primers P5 and P6 carry<br />
extensions that hybridize to the GFP cod<strong>in</strong>g sequence and primers P3 and P4 carry extensions that hybridize to a fragment carry<strong>in</strong>g the <strong>Aspergillus</strong><br />
fumigatus pyrG gene (AfpyrG) (Weidner et al., 1998). (B) After primer and nucleotide removal, these fragments were mixed with AfpyrG and gfp fragments<br />
and amplified with primers P2 and P7, which are nested primers. The five pieces hybridize dur<strong>in</strong>g fusion PCR and amplification results <strong>in</strong> a s<strong>in</strong>gle fragment<br />
as shown. Primers were designed such that GFP is fused <strong>in</strong> frame with the <strong>sumO</strong> cod<strong>in</strong>g sequence and the fusion is under control of the endogenous <strong>sumO</strong><br />
promoter. Upon transformation this fragment <strong>in</strong>tegrates at the <strong>sumO</strong> locus by homologous recomb<strong>in</strong>ation, replac<strong>in</strong>g the endogenous <strong>sumO</strong> gene with the<br />
construct shown. (For <strong>in</strong>terpretation of the references to colour <strong>in</strong> this figure legend, the reader is referred to the web version of this article.)