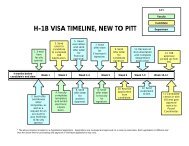
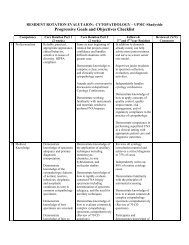
IDH1 and IDH2 Mutations Test Description: Detection of mutations in ...
IDH1 and IDH2 Mutations Test Description: Detection of mutations in ...
IDH1 and IDH2 Mutations Test Description: Detection of mutations in ...
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
<strong>IDH1</strong> <strong>and</strong> <strong>IDH2</strong> <strong>Mutations</strong><br />
<strong>Test</strong> <strong>Description</strong>:<br />
<strong>Detection</strong> <strong>of</strong> <strong>mutations</strong> <strong>in</strong> the <strong>IDH1</strong> <strong>and</strong> <strong>IDH2</strong> genes <strong>in</strong> bra<strong>in</strong> tumors.<br />
Indication:<br />
<strong>Mutations</strong> <strong>in</strong> either isocitrate dehydrogenase enzyme is<strong>of</strong>orms 1 (<strong>IDH1</strong>) <strong>and</strong> 2 (<strong>IDH2</strong>)<br />
genes have recently been identified <strong>in</strong> a large proportion <strong>of</strong> diffuse astrocytomas (70‐90%),<br />
oligodendrogliomas (69‐94%), oligoastrocytomas (78‐100%), <strong>and</strong> secondary<br />
glioblastomas (82‐88%)(1‐7). Primary glioblastomas showed low frequency <strong>of</strong> <strong>IDH1</strong>/<strong>IDH2</strong><br />
<strong>mutations</strong> (0‐5%). These <strong>mutations</strong> are not found <strong>in</strong> non‐neoplastic conditions that can<br />
<strong>of</strong>ten mimic gliomas (e.g. radiation changes, viral <strong>in</strong>fections, <strong>in</strong>farctions, etc.) (7).<br />
Approximately 90% <strong>of</strong> all <strong>mutations</strong> occur <strong>in</strong> codon 132, exon 4 <strong>of</strong> the <strong>IDH1</strong> gene, <strong>and</strong> less<br />
frequently <strong>in</strong> codon 172, exon 4 <strong>of</strong> the <strong>IDH2</strong> gene (4). It appears that patients with tumors<br />
harbor<strong>in</strong>g <strong>IDH1</strong> <strong>and</strong> <strong>IDH2</strong> <strong>mutations</strong> have better outcome than those with wild‐type IDH<br />
genes (4, 6).<br />
1. Balss J, Meyer J, Mueller W, et al. Analysis <strong>of</strong> the <strong>IDH1</strong> codon 132 mutation <strong>in</strong> bra<strong>in</strong><br />
tumors. Acta Neuropathol 2008:116;597‐602.<br />
2. Bleeker FE, Lamba S, Leenstra S, et al. <strong>IDH1</strong> <strong>mutations</strong> at residue p.R132<br />
(<strong>IDH1</strong>(R132)) occur frequently <strong>in</strong> high‐grade gliomas but not <strong>in</strong> other solid tumors.<br />
Hum Mutat 2009:30;7‐11.<br />
3. Parsons DW, Jones S, Zhang X, et al. An <strong>in</strong>tegrated genomic analysis <strong>of</strong> human<br />
glioblastoma multiforme. Science 2008:321;1807‐12.<br />
4. Yan H, Parsons DW, J<strong>in</strong> G, et al. <strong>IDH1</strong> <strong>and</strong> <strong>IDH2</strong> <strong>mutations</strong> <strong>in</strong> gliomas. N Engl J Med<br />
2009:360;765‐73.<br />
5. Kang MR, Kim MS, Oh JE, et al. Mutational analysis <strong>of</strong> <strong>IDH1</strong> codon 132 <strong>in</strong> glioblastomas<br />
<strong>and</strong> other common cancers. Int J Cancer 2009:125;353‐5.<br />
6. Sanson M, Marie Y, Paris S, et al. Isocitrate Dehydrogenase 1 Codon 132 Mutation Is an<br />
Important Prognostic Biomarker <strong>in</strong> Gliomas. J Cl<strong>in</strong> Oncol 2009.<br />
7. Horb<strong>in</strong>ski C, K<strong>of</strong>ler J, Kelly L, Murdoch GH, Nikiforova MN. The diagnostic utility <strong>of</strong><br />
<strong>IDH1</strong>/2 mutation analysis <strong>in</strong> rout<strong>in</strong>e cl<strong>in</strong>ical test<strong>in</strong>g <strong>of</strong> formal<strong>in</strong>‐fixed paraff<strong>in</strong>embedded<br />
glioma tissues. J Neuropathol Exp Neurol 2009 (<strong>in</strong> press).<br />
Methodology:<br />
H&E slides are reviewed by a pathologist to confirm the specimen adequacy <strong>and</strong> select an<br />
area <strong>of</strong> tumor for the molecular analysis. Tumor cells are manually microdissected for DNA<br />
isolation. Real time PCR <strong>and</strong> post‐PCR melt<strong>in</strong>g curve analysis is used for detection <strong>of</strong><br />
<strong>mutations</strong>. Positive results are confirmed by sequenc<strong>in</strong>g analysis.
Loci <strong>Test</strong>ed:<br />
<strong>IDH1</strong> <strong>and</strong> <strong>IDH2</strong><br />
Performed: Mon‐Fri<br />
Reported: With<strong>in</strong> 5 bus<strong>in</strong>ess days <strong>of</strong> receipt.<br />
Specimens Required:<br />
1 H&E <strong>and</strong> 6 blank slides or frozen tissue.<br />
Shipment Must Include:<br />
1 H&E <strong>and</strong> 6 blank slides <strong>of</strong> each block or frozen tissue<br />
MAP lab requisition form<br />
Patient’s pathology report