Isolation and identification of bacteria and fungi from ... - ictp
Isolation and identification of bacteria and fungi from ... - ictp
Isolation and identification of bacteria and fungi from ... - ictp
- No tags were found...
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
62<br />
ARTICLE IN PRESS<br />
C. Abrusci et al. / International Biodeterioration & Biodegradation 56 (2005) 58–68<br />
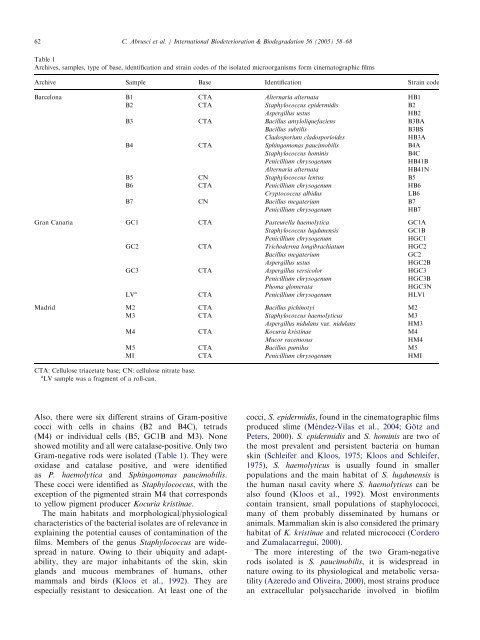
Table 1<br />
Archives, samples, type <strong>of</strong> base, <strong>identification</strong> <strong>and</strong> strain codes <strong>of</strong> the isolated microorganisms form cinematographic films<br />
Archive Sample Base Identification Strain code<br />
Barcelona B1 CTA Alternaria alternata HB1<br />
B2 CTA Staphylococcus epidermidis B2<br />
Aspergillus ustus<br />
HB2<br />
B3 CTA Bacillus amyloliquefaciens B3BA<br />
Bacillus subtilis<br />
B3BS<br />
Cladosporium cladosporioides<br />
HB3A<br />
B4 CTA Sphingomonas paucimobilis B4A<br />
Staphylococcus hominis<br />
B4C<br />
Penicillium chrysogenum<br />
HB41B<br />
Alternaria alternata<br />
HB41N<br />
B5 CN Staphylococcus lentus B5<br />
B6 CTA Penicillium chrysogenum HB6<br />
Cryptococcus albidus<br />
LB6<br />
B7 CN Bacillus megaterium B7<br />
Penicillium chrysogenum<br />
HB7<br />
Gran Canaria GC1 CTA Pasteurella haemolytica GC1A<br />
Staphylococcus lugdunensis<br />
GC1B<br />
Penicillium chrysogenum<br />
HGC1<br />
GC2 CTA Trichoderma longibrachiatum HGC2<br />
Bacillus megaterium<br />
GC2<br />
Aspergillus ustus<br />
HGC2B<br />
GC3 CTA Aspergillus versicolor HGC3<br />
Penicillium chrysogenum<br />
HGC3B<br />
Phoma glomerata<br />
HGC3N<br />
LV a CTA Penicillium chrysogenum HLV1<br />
Madrid M2 CTA Bacillus pichinotyi M2<br />
M3 CTA Staphylococcus haemolyticus M3<br />
Aspergillus nidulans var. nidulans<br />
HM3<br />
M4 CTA Kocuria kristinae M4<br />
Mucor racemosus<br />
HM4<br />
M5 CTA Bacillus pumilus M5<br />
MI CTA Penicillium chrysogenum HMI<br />
CTA: Cellulose triacetate base; CN: cellulose nitrate base.<br />
a LV sample was a fragment <strong>of</strong> a roll-can.<br />
Also, there were six different strains <strong>of</strong> Gram-positive<br />
cocci with cells in chains (B2 <strong>and</strong> B4C), tetrads<br />
(M4) or individual cells (B5, GC1B <strong>and</strong> M3).None<br />
showed motility <strong>and</strong> all were catalase-positive.Only two<br />
Gram-negative rods were isolated (Table 1).They were<br />
oxidase <strong>and</strong> catalase positive, <strong>and</strong> were identified<br />
as P. haemolytica <strong>and</strong> Sphingomonas paucimobilis.<br />
These cocci were identified as Staphylococcus, with the<br />
exception <strong>of</strong> the pigmented strain M4 that corresponds<br />
to yellow pigment producer Kocuria kristinae.<br />
The main habitats <strong>and</strong> morphological/physiological<br />
characteristics <strong>of</strong> the <strong>bacteria</strong>l isolates are <strong>of</strong> relevance in<br />
explaining the potential causes <strong>of</strong> contamination <strong>of</strong> the<br />
films.Members <strong>of</strong> the genus Staphylococcus are widespread<br />
in nature.Owing to their ubiquity <strong>and</strong> adaptability,<br />
they are major inhabitants <strong>of</strong> the skin, skin<br />
gl<strong>and</strong>s <strong>and</strong> mucous membranes <strong>of</strong> humans, other<br />
mammals <strong>and</strong> birds (Kloos et al., 1992).They are<br />
especially resistant to desiccation.At least one <strong>of</strong> the<br />
cocci, S. epidermidis, found in the cinematographic films<br />
produced slime (Me´ ndez-Vilas et al., 2004; Go¨ tz <strong>and</strong><br />
Peters, 2000). S. epidermidis <strong>and</strong> S. hominis are two <strong>of</strong><br />
the most prevalent <strong>and</strong> persistent <strong>bacteria</strong> on human<br />
skin (Schleifer <strong>and</strong> Kloos, 1975; Kloos <strong>and</strong> Schleifer,<br />
1975), S. haemolyticus is usually found in smaller<br />
populations <strong>and</strong> the main habitat <strong>of</strong> S. lugdunensis is<br />
the human nasal cavity where S. haemolyticus can be<br />
also found (Kloos et al., 1992).Most environments<br />
contain transient, small populations <strong>of</strong> staphylococci,<br />
many <strong>of</strong> them probably disseminated by humans or<br />
animals.Mammalian skin is also considered the primary<br />
habitat <strong>of</strong> K. kristinae <strong>and</strong> related micrococci (Cordero<br />
<strong>and</strong> Zumalacarregui, 2000).<br />
The more interesting <strong>of</strong> the two Gram-negative<br />
rods isolated is S. paucimobilis, it is widespread in<br />
nature owing to its physiological <strong>and</strong> metabolic versatility<br />
(Azeredo <strong>and</strong> Oliveira, 2000), most strains produce<br />
an extracellular polysaccharide involved in bi<strong>of</strong>ilm