icvg 2009 part I pp 1-131.pdf - Cornell University
icvg 2009 part I pp 1-131.pdf - Cornell University
icvg 2009 part I pp 1-131.pdf - Cornell University
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
— 25 —<br />
immunoassays, either ELISA, ISEM, IPEM or Western<br />
blots. They can discriminate unequivocally between related<br />
viruses or variants in samples from mixed infections. As the<br />
evolution of viruses is often paralleled by changes of the<br />
coat protein, Mabs might well distinguish biological<br />
distinct variants. Mabs may be used to map epitopes on<br />
viral coat proteins (Zhou et al., 2003) or to <strong>part</strong>ially<br />
decorate virus <strong>part</strong>icles (Gugerli et al., 1993). They can<br />
easily be mixed with others to make efficient diagnostic<br />
tools. The development of Mabs to GLRaVs can however<br />
be laborious and costly. In some circumstances, Mabs may<br />
be too specific. This was eventually leading to an<br />
unnecessary segregation of provisional virus species, as in<br />
the case of GLRaV-4 or GLRaV-5 related viruses. As<br />
mentioned above, these disadvantages can at the same time<br />
be positive. As an example, specific Mabs to GLRaV-6<br />
permitted to trace the intriguing massive occurrence of this<br />
virus species in the cultivar Cardinal (Boscia et al., 2000).<br />
Finally, some Mabs are difficult to handle.<br />
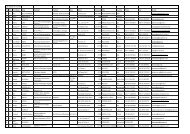
Table 1. Production of antiserum (As), monoclonal antibodies (Mabs) and recombinant antibodies (recAbs) to Grapevine leafroll-associated<br />
viruses (GLRaVs)<br />
Antibody Immunogen Cultivar Reference<br />
As; Mab 2-4 GLRaV-1 Räuschling 24 CH Gugerli et al., 1984, 1987<br />
As GLRaV-2 Gamay RdL 356/r.37 CH Rosciglione et al., 1986<br />
As; Mab 8 GLRaV-3 Fra<strong>pp</strong>ato Mortillo I Gugerli et al., 1987, 1990<br />
As GLRaV-1 Chardonnay F Legin et al., 1987<br />
As; Mab GLRaV-3 Pinot noir NY-1 USA Zee et al., 1987; Madden et al., 1987<br />
As GLRaV-1 Klevner Heiligenstein F Zimmermann et al., 1988<br />
Mabs NY1.1 to 1.4 GLRaV-3 Pinot noir NY-1 USA Hu et al., 1989, 1990a, 1991<br />
As GLRaV-2 Chaouch Rose TR Zimmermann et al., 1990a<br />
As; Mabs 1 to 3 GLRaV-3 Chardonnay F Zimmermann et al., 1990a, 1990b<br />
As GLRaV-5 1) White Emperor USA Zimmermann et al., 1990a<br />
As GLRaV-4 Thompson USA Hu et al., 1990b<br />
As GLRaV-2+3 Crouchen 2/1/20 ZA Engelbrecht et al., 1990<br />
As; Mabs GLRaV-3 16G isolate I Faggioli F et al., 1991<br />
Mab 29-1, 29-2 GLRaV-2 Chasselas 8/22 CH Gugerli et al., 1993<br />
As; Mab 3-1, 6-3, 15-5 GLRaV-4 Thompson Seedless V.C.A2v22 USA Gugerli et al., 1993, Besse et al., <strong>2009</strong>b<br />
As; Mab 43-1, 3-3, 8-2 GLRaV-5 Emperor V.C.A2v18 - White USA Gugerli et al., 1993, Besse et al., <strong>2009</strong><br />
As GLRaV-1 to 5 CP Black Spanish 90/246 ZA Goszczynski et al., 1995<br />
As GLRaV-7 AA42 unidentified AL Choueiri et al., 1996<br />
As GLRaV-2 Muscat of Alexandria and N33 ZA Goszczynski et al., 1996a, 1996b<br />
As GLRaV-3 Zweigelt (Kecskemét) H Tobias et al., 1996<br />
As; Mab 36-117 GLRaV-2+6 Chasselas 8/22 CH Gugerli et al.1997<br />
As GLRaV-3 recCP Pinot noir NY-1 USA Ling et al., 1997, 2000<br />
As; Mabs 3F76, 14F9, 15F1, 19A12 GLRaV-8 Thompson Seedless LR102 USA Monis et al., 1997, 2000<br />
As GLRaV-1 CP French Colombart FC/2 USA Monis et al., 1997<br />
As GLRaV-2 CP Malbec (incompatibility) USA Monis et al., 1997<br />
As GLRaV-2 recCP N. benthamiana H4 USA Zhu et al., 1997<br />
As; Mabs 1G10 3) ,1B7 4) , GLRaV-1 Houedi Y233 (F) 2) Seddas et al., 2000<br />
1C4 5) , 2F11, 2F3 6)<br />
As GLRV-3 Raziki Y285 (F) 2) Seddas et al., 2000<br />
Mab R19 GLRaV-2 N. benthamiana H4 (I) 2) Zhou et al., 2000<br />
RecAb GLRaV-3 Nölke et al., 2003<br />
Mabs Nig.A, B, C and.I GLRaV-3 Moscato giallo NIG3 I Zhou et al., 2003a, 2003b<br />
As GLRaV-2 Y252 IL Saldarelli et al., 2006<br />
As GLRaV-2 Y253 TR Saldarelli et al., 2006<br />
As ; Mabs 6-5, 37-15 GLRaV-7 Y276 (F) 2) Rigotti et al., 2006<br />
As GLRaV-2 recCP Cabernet Franc CN Xu et al., 2006<br />
As GLRaV-2 recCP Pinot noir USA Ling et al., 2007<br />
As; Mab 5A5/C2, 8G5/H6 GLRaV-3 recCP Merlot Cl-766 RCH Engel et al., 2008<br />
RecAb scFvLR3cp-1 GLRaV-3 (I) 2) Orecchia et al., 2008<br />
As GLRaV-2 recCP LN33 BR Radaelli et al., 2008<br />
RecAb C L -LR3 GLRaV-3 (I) 2) Cogotzi et al., <strong>2009</strong><br />
As 1295 ; Mab 4-2 , Mab 8-2-3 GLRaV-2 variant Pinot noir 20/50 CH Besse et al., <strong>2009</strong>a<br />
As; Mabs 62-4, 27-1 GLRaV-9 Cabernet Sauvignon AUS Gugerli et al., <strong>2009</strong><br />
As GLRaV-Pr recCP Prevezaniko GR Maliogka et al., <strong>2009</strong><br />
1) Originally named GLRaV-4; 2) Origin not stated; from collection (F) INRA Colmar or (I) <strong>University</strong> of Bari, Italia; 3) Cross-reacting with GLRaV-3; 4) Wide<br />
spectrum (32 out of 33 isolates tested); 5) Limited spectrum (25 out of 33 isolates tested); 6) Useful in diagnostic a<strong>pp</strong>lications; RecCP: recombinant coat<br />
protein; As : polyclonal antibody (mostly from immunised rabbits); CP : coat protein; RecAb : recombinant antibody made from single-chain variable<br />
Fragment (scFv)<br />
.<br />
REC-CP AND REC-AB<br />
The use of recombinant coat proteins as immunogen<br />
(Ling et al., 1997, 2000; Zhu et al., 1997; Xu et al., 2006;<br />
Ling et al 2007; Engel et al., 2008; Radaelli et al., 2008)<br />
and in vitro engineered recombinant antibodies (rAb) with<br />
single-chain variable fragment (scFv) (Nölke et al., 2003;<br />
Orecchia et al., 2008; Cogotzi et al., <strong>2009</strong>) reflect the<br />
development of exciting new technologies.<br />
Progrès Agricole et Viticole, <strong>2009</strong>, Hors Série – Extended abstracts 16 th Meeting of ICVG, Dijon, France, 31 Aug – 4 Sept <strong>2009</strong>