4 Multiple Sequence Alignment 4.1 Multiple sequence alignment
4 Multiple Sequence Alignment 4.1 Multiple sequence alignment
4 Multiple Sequence Alignment 4.1 Multiple sequence alignment
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
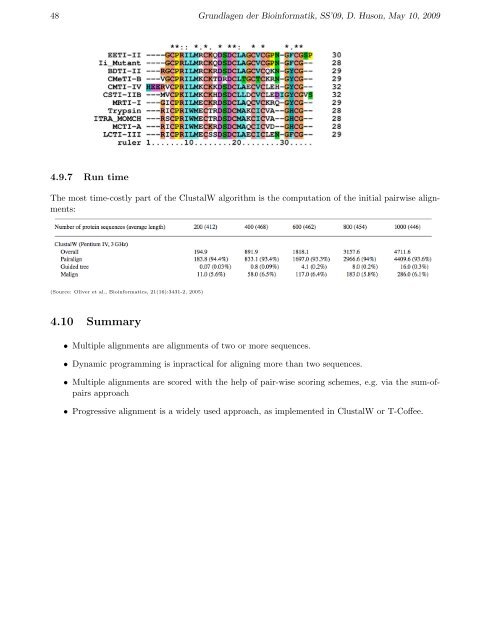
48 Grundlagen der Bioinformatik, SS’09, D. Huson, May 10, 20094.9.7 Run timeThe most time-costly part of the ClustalW algorithm is the computation of the initial pairwise <strong>alignment</strong>s:(Source: Oliver et al., Bioinformatics, 21(16):3431-2, 2005)<strong>4.1</strong>0 Summary• <strong>Multiple</strong> <strong>alignment</strong>s are <strong>alignment</strong>s of two or more <strong>sequence</strong>s.• Dynamic programming is inpractical for aligning more than two <strong>sequence</strong>s.• <strong>Multiple</strong> <strong>alignment</strong>s are scored with the help of pair-wise scoring schemes, e.g. via the sum-ofpairsapproach• Progressive <strong>alignment</strong> is a widely used approach, as implemented in ClustalW or T-Coffee.