4 Multiple Sequence Alignment 4.1 Multiple sequence alignment
4 Multiple Sequence Alignment 4.1 Multiple sequence alignment
4 Multiple Sequence Alignment 4.1 Multiple sequence alignment
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
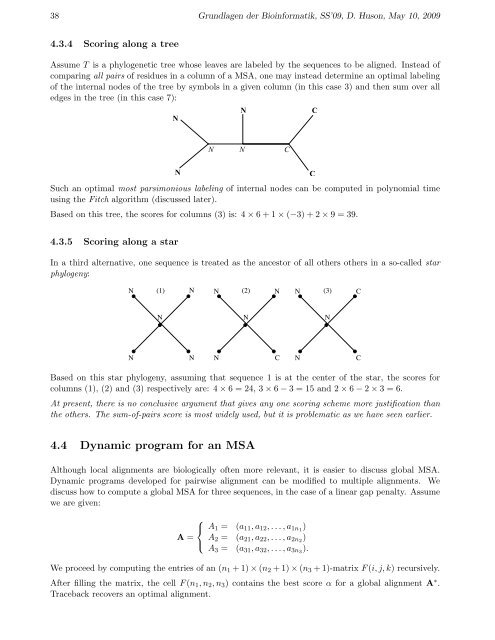
38 Grundlagen der Bioinformatik, SS’09, D. Huson, May 10, 20094.3.4 Scoring along a treeAssume T is a phylogenetic tree whose leaves are labeled by the <strong>sequence</strong>s to be aligned. Instead ofcomparing all pairs of residues in a column of a MSA, one may instead determine an optimal labelingof the internal nodes of the tree by symbols in a given column (in this case 3) and then sum over alledges in the tree (in this case 7):NNCNNCNSuch an optimal most parsimonious labeling of internal nodes can be computed in polynomial timeusing the Fitch algorithm (discussed later).Based on this tree, the scores for columns (3) is: 4 × 6 + 1 × (−3) + 2 × 9 = 39.C4.3.5 Scoring along a starIn a third alternative, one <strong>sequence</strong> is treated as the ancestor of all others others in a so-called starphylogeny:N(1) N N (2) N N (3)CNNNNNNCNCBased on this star phylogeny, assuming that <strong>sequence</strong> 1 is at the center of the star, the scores forcolumns (1), (2) and (3) respectively are: 4 × 6 = 24, 3 × 6 − 3 = 15 and 2 × 6 − 2 × 3 = 6.At present, there is no conclusive argument that gives any one scoring scheme more justification thanthe others. The sum-of-pairs score is most widely used, but it is problematic as we have seen earlier.4.4 Dynamic program for an MSAAlthough local <strong>alignment</strong>s are biologically often more relevant, it is easier to discuss global MSA.Dynamic programs developed for pairwise <strong>alignment</strong> can be modified to multiple <strong>alignment</strong>s. Wediscuss how to compute a global MSA for three <strong>sequence</strong>s, in the case of a linear gap penalty. Assumewe are given:⎧⎨ A 1 = (a 11 , a 12 , . . . , a 1n1 )A = A 2 =⎩A 3 =(a 21 , a 22 , . . . , a 2n2 )(a 31 , a 32 , . . . , a 3n3 ).We proceed by computing the entries of an (n 1 + 1) × (n 2 + 1) × (n 3 + 1)-matrix F (i, j, k) recursively.After filling the matrix, the cell F (n 1 , n 2 , n 3 ) contains the best score α for a global <strong>alignment</strong> A ∗ .Traceback recovers an optimal <strong>alignment</strong>.