Molecular Modeling & Simulation - Nano Mahidol - Mahidol University
Molecular Modeling & Simulation - Nano Mahidol - Mahidol University
Molecular Modeling & Simulation - Nano Mahidol - Mahidol University
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
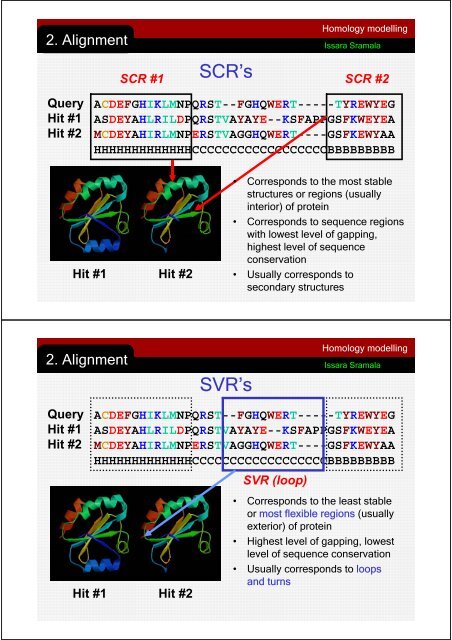
Homology modelling2. AlignmentIssara SramalaSCR #1SCR’sSCR #2QueryHit #1Hit #2ACDEFGHIKLMNPQRST--FGHQWERT-----TYREWYEGASDEYAHLRILDPQRSTVAYAYE--KSFAPPGSFKWEYEAMCDEYAHIRLMNPERSTVAGGHQWERT----GSFKEWYAAHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCBBBBBBBBBHit #1 Hit #2• Corresponds to the most stablestructures or regions (usuallyinterior) of protein• Corresponds to sequence regionswith lowest level of gapping,highest level of sequenceconservation• Usually corresponds tosecondary structuresQueryHit #1Hit #2SVR’sACDEFGHIKLMNPQRST--FGHQWERT-----TYREWYEGASDEYAHLRILDPQRSTVAYAYE--KSFAPPGSFKWEYEAMCDEYAHIRLMNPERSTVAGGHQWERT----GSFKEWYAAHHHHHHHHHHHHHCCCCCCCCCCCCCCCCCCBBBBBBBBBHit #1 Hit #2SVR (loop)Homology modelling2. AlignmentIssara Sramala• Corresponds to the least stableor most flexible regions (usuallyexterior) of protein• Highest level of gapping, lowestlevel of sequence conservation• Usually corresponds to loopsand turns