The utility of song and morphological characters in delineating ...
The utility of song and morphological characters in delineating ...
The utility of song and morphological characters in delineating ...
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
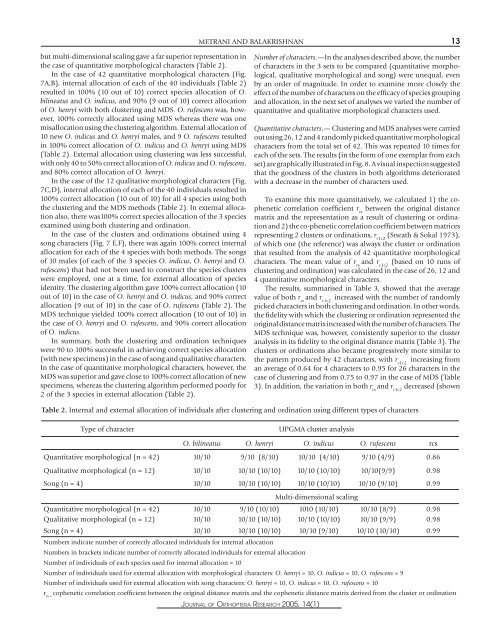
METRANI AND BALAKRISHNAN 13but multi-dimensional scal<strong>in</strong>g gave a far superior representation <strong>in</strong>the case <strong>of</strong> quantitative <strong>morphological</strong> <strong>characters</strong> (Table 2).In the case <strong>of</strong> 42 quantitative <strong>morphological</strong> <strong>characters</strong> (Fig.7A,B), <strong>in</strong>ternal allocation <strong>of</strong> each <strong>of</strong> the 40 <strong>in</strong>dividuals (Table 2)resulted <strong>in</strong> 100% (10 out <strong>of</strong> 10) correct species allocation <strong>of</strong> O.bil<strong>in</strong>eatus <strong>and</strong> O. <strong>in</strong>dicus, <strong>and</strong> 90% (9 out <strong>of</strong> 10) correct allocation<strong>of</strong> O. henryi with both cluster<strong>in</strong>g <strong>and</strong> MDS. O. rufescens was, however,100% correctly allocated us<strong>in</strong>g MDS whereas there was onemisallocation us<strong>in</strong>g the cluster<strong>in</strong>g algorithm. External allocation <strong>of</strong>10 new O. <strong>in</strong>dicus <strong>and</strong> O. henryi males, <strong>and</strong> 9 O. rufescens resulted<strong>in</strong> 100% correct allocation <strong>of</strong> O. <strong>in</strong>dicus <strong>and</strong> O. henryi us<strong>in</strong>g MDS(Table 2). External allocation us<strong>in</strong>g cluster<strong>in</strong>g was less successful,with only 40 to 50% correct allocation <strong>of</strong> O. <strong>in</strong>dicus <strong>and</strong> O. rufescens,<strong>and</strong> 80% correct allocation <strong>of</strong> O. henryi.In the case <strong>of</strong> the 12 qualitative <strong>morphological</strong> <strong>characters</strong> (Fig.7C,D), <strong>in</strong>ternal allocation <strong>of</strong> each <strong>of</strong> the 40 <strong>in</strong>dividuals resulted <strong>in</strong>100% correct allocation (10 out <strong>of</strong> 10) for all 4 species us<strong>in</strong>g boththe cluster<strong>in</strong>g <strong>and</strong> the MDS methods (Table 2). In external allocationalso, there was100% correct species allocation <strong>of</strong> the 3 speciesexam<strong>in</strong>ed us<strong>in</strong>g both cluster<strong>in</strong>g <strong>and</strong> ord<strong>in</strong>ation.In the case <strong>of</strong> the clusters <strong>and</strong> ord<strong>in</strong>ations obta<strong>in</strong>ed us<strong>in</strong>g 4<strong>song</strong> <strong>characters</strong> (Fig. 7 E,F), there was aga<strong>in</strong> 100% correct <strong>in</strong>ternalallocation for each <strong>of</strong> the 4 species with both methods. <strong>The</strong> <strong>song</strong>s<strong>of</strong> 10 males (<strong>of</strong> each <strong>of</strong> the 3 species O. <strong>in</strong>dicus, O. henryi <strong>and</strong> O.rufescens) that had not been used to construct the species clusterswere employed, one at a time, for external allocation <strong>of</strong> speciesidentity. <strong>The</strong> cluster<strong>in</strong>g algorithm gave 100% correct allocation (10out <strong>of</strong> 10) <strong>in</strong> the case <strong>of</strong> O. henryi <strong>and</strong> O. <strong>in</strong>dicus, <strong>and</strong> 90% correctallocation (9 out <strong>of</strong> 10) <strong>in</strong> the case <strong>of</strong> O. rufescens (Table 2). <strong>The</strong>MDS technique yielded 100% correct allocation (10 out <strong>of</strong> 10) <strong>in</strong>the case <strong>of</strong> O. henryi <strong>and</strong> O. rufescens, <strong>and</strong> 90% correct allocation<strong>of</strong> O. <strong>in</strong>dicus.In summary, both the cluster<strong>in</strong>g <strong>and</strong> ord<strong>in</strong>ation techniqueswere 90 to 100% successful <strong>in</strong> achiev<strong>in</strong>g correct species allocation(with new specimens) <strong>in</strong> the case <strong>of</strong> <strong>song</strong> <strong>and</strong> qualitative <strong>characters</strong>.In the case <strong>of</strong> quantitative <strong>morphological</strong> <strong>characters</strong>, however, theMDS was superior <strong>and</strong> gave close to 100% correct allocation <strong>of</strong> newspecimens, whereas the cluster<strong>in</strong>g algorithm performed poorly for2 <strong>of</strong> the 3 species <strong>in</strong> external allocation (Table 2).Number <strong>of</strong> <strong>characters</strong>.—In the analyses described above, the number<strong>of</strong> <strong>characters</strong> <strong>in</strong> the 3 sets to be compared (quantitative <strong>morphological</strong>,qualitative <strong>morphological</strong> <strong>and</strong> <strong>song</strong>) were unequal, evenby an order <strong>of</strong> magnitude. In order to exam<strong>in</strong>e more closely theeffect <strong>of</strong> the number <strong>of</strong> <strong>characters</strong> on the efficacy <strong>of</strong> species group<strong>in</strong>g<strong>and</strong> allocation, <strong>in</strong> the next set <strong>of</strong> analyses we varied the number <strong>of</strong>quantitative <strong>and</strong> qualitative <strong>morphological</strong> <strong>characters</strong> used.Quantitative <strong>characters</strong>.— Cluster<strong>in</strong>g <strong>and</strong> MDS analyses were carriedout us<strong>in</strong>g 26, 12 <strong>and</strong> 4 r<strong>and</strong>omly picked quantitative <strong>morphological</strong><strong>characters</strong> from the total set <strong>of</strong> 42. This was repeated 10 times foreach <strong>of</strong> the sets. <strong>The</strong> results (<strong>in</strong> the form <strong>of</strong> one exemplar from eachset) are graphically illustrated <strong>in</strong> Fig. 8. A visual <strong>in</strong>spection suggestedthat the goodness <strong>of</strong> the clusters <strong>in</strong> both algorithms deterioratedwith a decrease <strong>in</strong> the number <strong>of</strong> <strong>characters</strong> used.To exam<strong>in</strong>e this more quantitatively, we calculated 1) the copheneticcorrelation coefficient r csbetween the orig<strong>in</strong>al distancematrix <strong>and</strong> the representation as a result <strong>of</strong> cluster<strong>in</strong>g or ord<strong>in</strong>ation<strong>and</strong> 2) the co-phenetic correlation coefficient between matricesrepresent<strong>in</strong>g 2 clusters or ord<strong>in</strong>ations, r c1c2(Sneath & Sokal 1973),<strong>of</strong> which one (the reference) was always the cluster or ord<strong>in</strong>ationthat resulted from the analysis <strong>of</strong> 42 quantitative <strong>morphological</strong><strong>characters</strong>. <strong>The</strong> mean value <strong>of</strong> r cs<strong>and</strong> r c1c2(based on 10 runs <strong>of</strong>cluster<strong>in</strong>g <strong>and</strong> ord<strong>in</strong>ation) was calculated <strong>in</strong> the case <strong>of</strong> 26, 12 <strong>and</strong>4 quantitative <strong>morphological</strong> <strong>characters</strong>.<strong>The</strong> results, summarised <strong>in</strong> Table 3, showed that the averagevalue <strong>of</strong> both r cs<strong>and</strong> r <strong>in</strong>creased with the number <strong>of</strong> r<strong>and</strong>omlyc1c2picked <strong>characters</strong> <strong>in</strong> both cluster<strong>in</strong>g <strong>and</strong> ord<strong>in</strong>ation. In other words,the fidelity with which the cluster<strong>in</strong>g or ord<strong>in</strong>ation represented theorig<strong>in</strong>al distance matrix <strong>in</strong>creased with the number <strong>of</strong> <strong>characters</strong>. <strong>The</strong>MDS technique was, however, consistently superior to the clusteranalysis <strong>in</strong> its fidelity to the orig<strong>in</strong>al distance matrix (Table 3). <strong>The</strong>clusters or ord<strong>in</strong>ations also became progressively more similar tothe pattern produced by 42 <strong>characters</strong>, with r c1c2<strong>in</strong>creas<strong>in</strong>g froman average <strong>of</strong> 0.64 for 4 <strong>characters</strong> to 0.95 for 26 <strong>characters</strong> <strong>in</strong> thecase <strong>of</strong> cluster<strong>in</strong>g <strong>and</strong> from 0.75 to 0.97 <strong>in</strong> the case <strong>of</strong> MDS (Table3). In addition, the variation <strong>in</strong> both r cs<strong>and</strong> r c1c2decreased (shownTable 2. Internal <strong>and</strong> external allocation <strong>of</strong> <strong>in</strong>dividuals after cluster<strong>in</strong>g <strong>and</strong> ord<strong>in</strong>ation us<strong>in</strong>g different types <strong>of</strong> <strong>characters</strong>Type <strong>of</strong> characterUPGMA cluster analysisO. bil<strong>in</strong>eatus O. henryi O. <strong>in</strong>dicus O. rufescens rcsQuantitative <strong>morphological</strong> (n = 42) 10/10 9/10 (8/10) 10/10 (4/10) 9/10 (4/9) 0.86Qualitative <strong>morphological</strong> (n = 12) 10/10 10/10 (10/10) 10/10 (10/10) 10/10(9/9) 0.98Song (n = 4) 10/10 10/10 (10/10) 10/10 (10/10) 10/10 (9/10) 0.99Multi-dimensional scal<strong>in</strong>gQuantitative <strong>morphological</strong> (n = 42) 10/10 9/10 (10/10) 1010 (10/10) 10/10 (8/9) 0.98Qualitative <strong>morphological</strong> (n = 12) 10/10 10/10 (10/10) 10/10 (10/10) 10/10 (9/9) 0.98Song (n = 4) 10/10 10/10 (10/10) 10/10 (9/10) 10/10 (10/10) 0.99Numbers <strong>in</strong>dicate number <strong>of</strong> correctly allocated <strong>in</strong>dividuals for <strong>in</strong>ternal allocationNumbers <strong>in</strong> brackets <strong>in</strong>dicate number <strong>of</strong> correctly allocated <strong>in</strong>dividuals for external allocationNumber <strong>of</strong> <strong>in</strong>dividuals <strong>of</strong> each species used for <strong>in</strong>ternal allocation = 10Number <strong>of</strong> <strong>in</strong>dividuals used for external allocation with <strong>morphological</strong> <strong>characters</strong>: O. henryi = 10, O. <strong>in</strong>dicus = 10, O. rufescens = 9Number <strong>of</strong> <strong>in</strong>dividuals used for external allocation with <strong>song</strong> <strong>characters</strong>: O. henryi = 10, O. <strong>in</strong>dicus = 10, O. rufescens = 10r cs =cophenetic correlation coefficient between the orig<strong>in</strong>al distance matrix <strong>and</strong> the cophenetic distance matrix derived from the cluster or ord<strong>in</strong>ationJOURNAL OF ORTHOPTERA RESEARCH 2005, 14(1)