Genetic Molecular Techiniques - Part V - RNAi, Human Disease
Genetic Molecular Techiniques - Part V - RNAi, Human Disease
Genetic Molecular Techiniques - Part V - RNAi, Human Disease
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
Amy Pasquinelli, Office Phone: 858-822-3006<br />
apasquin@ucsd.edu, Subject: BIMM100<br />
Wed., April 28, 5-6PM, Bonner Hall 4146<br />
MIDTERM EXAM - TUESDAY, MAY 4<br />
6:30-8PM, SOLIS 107 (this room and time)<br />
Emergencies regarding exam - Submit in writing + full<br />
documentation. Make-up is oral exam by Dr. Niwa.<br />
You need a pen for the exam and nothing else -<br />
personals can be kept in the front of the room.<br />
There is ZERO tolerance for cheating.<br />
Exams will be returned when Dr. Niwa gets back.<br />
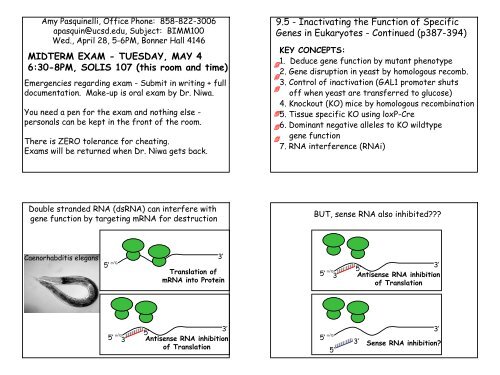
Double stranded RNA (dsRNA) can interfere with<br />
gene function by targeting mRNA for destruction<br />
Caenorhabditis elegans<br />
m7G 5’<br />
5’<br />
m 7 G<br />
3’<br />
3’<br />
Translation of<br />
mRNA into Protein<br />
3’<br />
5’<br />
Antisense RNA inhibition<br />
of Translation<br />
9.5 - Inactivating the Function of Specific<br />
Genes in Eukaryotes - Continued (p387-394)<br />
�<br />
�<br />
�<br />
�<br />
�<br />
�<br />
KEY CONCEPTS:<br />
1. Deduce gene function by mutant phenotype<br />
2. Gene disruption in yeast by homologous recomb.<br />
3. Control of inactivation (GAL1 promoter shuts<br />
off when yeast are transferred to glucose)<br />
4. Knockout (KO) mice by homologous recombination<br />
5. Tissue specific KO using loxP-Cre<br />
6. Dominant negative alleles to KO wildtype<br />
gene function<br />
7. RNA interference (<strong>RNAi</strong>)<br />
BUT, sense RNA also inhibited???<br />
5’<br />
m 7 G<br />
m7G 5’<br />
3’<br />
5’<br />
3’<br />
5’<br />
Antisense RNA inhibition<br />
of Translation<br />
3’<br />
3’<br />
Sense RNA inhibition?
The Solution: dsRNA directs mRNA Degradation!<br />
input dsRNA<br />
in vitro transcription<br />
of dsRNA<br />
RNA interference (<strong>RNAi</strong>)<br />
processing<br />
RNase<br />
~22nt siRNAs<br />
5’ 3’<br />
3’ 5’<br />
target<br />
recognition<br />
mRNA<br />
5’ 3’<br />
3’ 5’<br />
degradation<br />
Fig. 9-43a<br />
injection of dsRNA<br />
degradation of target<br />
mRNA in progeny - “KO”<br />
-specific<br />
-almost any mRNA can be<br />
targeted<br />
-defense against RNA viruses<br />
-works in human cells<br />
-high throughput screening<br />
of gene function with <strong>RNAi</strong><br />
“libraries”<br />
Analysis of Gene Function by <strong>RNAi</strong> KO<br />
1. Synthesize dsRNA in vitro<br />
2. Inject parent worm with dsRNA<br />
3. Test effect on mRNA expression by in situ<br />
4. What is the phenotype?<br />
Fig. 9-43b<br />
mex-3 mRNA present mex-3 mRNA absent<br />
9.6 Identifying and Locating <strong>Human</strong> <strong>Disease</strong> Genes<br />
KEY CONCEPTS:<br />
1. Patterns of inheritance: Autosomal dom.,<br />
Autosomal recessive, X-linked rec.<br />
2. Recombinant genotypes by crossing over<br />
3. Mapping - cosegregation with markers<br />
4. <strong>Molecular</strong> markers - polymorphisms<br />
5. RFLPs, SNPs, SSRs<br />
6. Expression analysis, DNA sequencing<br />
7. <strong>Genetic</strong> heterogeneity and polygenic traits
3 Major Patterns of <strong>Disease</strong> Inheritance<br />
1. Autosomal dominant<br />
- at least one parent is a carrier<br />
- usually post-reproductive<br />
- Huntington’s (1 mutant HD allele)<br />
2. Autosomal recessive<br />
- both parents must be carriers<br />
- cystic fibrosis (defective chloride channel<br />
gene (CFTR)<br />
3. X-linked recessive<br />
- mother is carrier on X chromosome<br />
- males get X from mom - 50% chance of mutant<br />
allele (X M /Y)<br />
- Duchenne muscular dystrophy (DMD)<br />
3 Major Patterns of <strong>Disease</strong> Inheritance<br />
25%<br />
50%<br />
50%<br />
How do you find the disease causing gene?<br />
Recombinational analysis<br />
Fig. 9-45<br />
1. pairs of homologous<br />
chromo. align<br />
2. crossing over<br />
3. the closer the genes,<br />
the less frequent<br />
they will crossover,<br />
i.e. the more tightly<br />
they are linked<br />
The ordering of genes along a chromosome is a genetic map, or linkage map
DNA Polymorphisms - variations in DNA sequence<br />
RFLPs - restriction fragment length polymorphisms<br />
SNPs - single nucleotide polymorphisms<br />
SSRs - simple sequence repeats or microsatellites<br />
Pedigree based on RFLP analysis<br />
Fig. 9-46b<br />
Experimental Detection of RFLPs<br />
WT<br />
M<br />
Fig. 9-46a<br />
How do you find the disease causing gene (cont.)?<br />
1. Comparison of gene expression from genomic region<br />
a. Northern blotting<br />
b. in situs<br />
2. Sequence candidate genes from region<br />
Example: Sickle Cell Anemia<br />
Chromosome 11<br />
collagen<br />
ribosome<br />
hemoglobin<br />
iron transporter<br />
LOC139168
Many <strong>Disease</strong> genes result from<br />
multiple genetic defects<br />
1. Monogenetic traits - defect in 1 gene<br />
Examples - see Table 9-3<br />
2. <strong>Genetic</strong> heterogeneity - defects in a variety<br />
of different genes produce same disease<br />
Example - Retinitis pigmentosa can be caused<br />
by mutation in more than 60 genes!<br />
3. Polygenic traits - defects in multiple<br />
different genes contribute to the disease<br />
Examples - diabetes, heart disease...<br />
GOOD LUCK STUDYING<br />
FOR YOUR EXAM!