Complete issue - IMA Fungus
Complete issue - IMA Fungus
Complete issue - IMA Fungus
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
T h e G l o b a l<br />
M y c o l o g i c a l J o u r n a l<br />
Volume 2 · No. 2 · December 2011<br />
N E W S · R E P O R T S · A W A R D S A N D P E R S O N A L I A · R E S E A R C H N E W S<br />
B O O K N E W S · f o r t h c o m i n g M E E T I N G S · A R T I C L E S
Colofon<br />
<strong>IMA</strong> <strong>Fungus</strong><br />
Compiled by the International<br />
Mycological Association for the<br />
world’s mycologists.<br />
Scope: All aspects of pure and<br />
applied mycological research and<br />
news.<br />
Aims: To be the flagship journal<br />
of the International Mycological<br />
Association. <strong>IMA</strong> FUNGUS is<br />
an international, peer-reviewed,<br />
open-access, full colour, fast-track<br />
journal.<br />
Frequency: Published twice per year<br />
(June and December). Articles are<br />
published online with final pagination<br />
as soon as they have been<br />
accepted and edited.<br />
ISSN<br />
E-ISSN<br />
2210-6340 (print)<br />
2210-6359 (online)<br />
Websites: www. imafungus.org<br />
www.ima-mycology.org<br />
E-mail: d.hawksworth@nhm.ac.uk<br />
Volume 2 · No. 2 · December 2011<br />
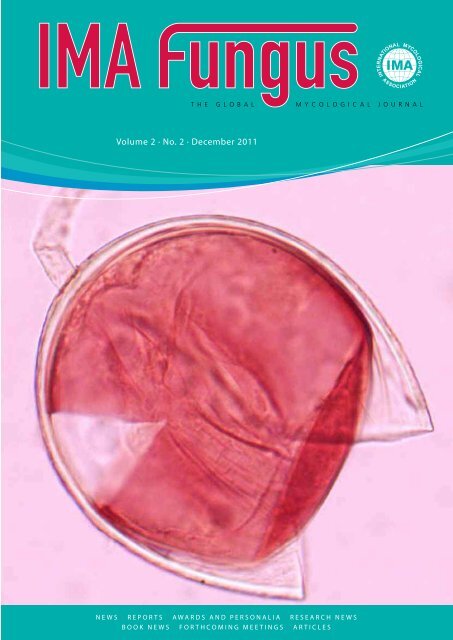
Cover: Crushed, bi-walled spore of<br />
Pacispora franciscana (ZT Myc)<br />
extracted from the type locality<br />
from mycorrhizospheric soil under<br />
an olive tree close to the Basilica of<br />
St Francis of Assisi (Umbria, Italy).<br />
The inner, germinal wall of the brilliant<br />
white spore stains in Melzer’s<br />
reagent. Photo Fritz Oehl.<br />
EDITORIAL BOARD<br />
Editor-in-Chief<br />
Prof. dr D.L. Hawksworth CBE, Departamento de Biología Vegetal II, Facultad de Farmacia, Universidad Complutense de<br />
Madrid, Plaza Ramón y Cajal, 28040 Madrid, Spain; and Department of Botany, Natural History Museum, Cromwell<br />
Road, London SW7 5BD, UK; E-mail: d.hawksworth@nhm.ac.uk<br />
Layout Editors<br />
M.J. van den Hoeven-Verweij & M. Vermaas, CBS-KNAW Fungal Biodiversity Centre, P.O. Box 85167, 3508 AD<br />
Utrecht, The Netherlands; E-mail: m.verweij@cbs.knaw.nl<br />
Associate Editors<br />
Dr T.V. Andrianova, M.G. Kholodny Institute of Botany, Tereshchenkivska Street 2, Kiev, MSP-1, 01601, Ukraine;<br />
E-mail: tand@darwin.relc.com<br />
Prof. dr D. Begerow, Lehrstuhl für Evolution und Biodiversität der Pflanzen, Ruhr-Universität Bochum, Universitätsstr.<br />
150, Gebäude ND 44780, Bochum, Germany; E-mail: dominik.begerow@rub.de<br />
Dr S. Cantrell, Department of Plant Pathology and Crop Physiology, Louisiana State University, Agricultural Centre, 455 Life<br />
Sciences Bldg., Baton Rouge, LA 70803, USA; E-mail: scantrel@suagm.edu<br />
Prof. dr D. Carter, Discipline of Microbiology, School of Molecular Biosciences, Building G08, University of Sydney,<br />
NSW 2006, Australia; E-mail: d.carter@mmb.usyd.edu.au<br />
Prof. dr P.W. Crous, CBS-KNAW Fungal Biodiversity Centre, P.O. Box 85167, 3508 AD Utrecht, The Netherlands; E-<br />
mail: p.crous@cbs.knaw.nl<br />
Prof. dr J. Dianese, Departamento de Fitopatologia, Universidade de Brasília, 70910-900 Brasília, D.F., Brasil; E-mail:<br />
jcarmine@unb.br<br />
Dr P.S. Dyer, School of Biology, Institute of Genetics, University of Nottingham, University Park, Nottingham NG7 2RD,<br />
UK; E-mail: paul.dyer@nottingham.ac.uk<br />
Dr M. Gryzenhout, Dept. of Plant Sciences, University of the Free State, P.O. Box 339, Bloemfontein 9300, South Africa;<br />
E-mail: Gryzenhoutm@ufs.ac.za<br />
Prof. dr L. Guzman-Davalos, Instituto de Botánica, Departamento de Botánica y Zoología, Universidad de Guadalajara,<br />
A.P. 1-139 Zapopan, 45101, México; E-mail: lguzman@cucba.udg.mx<br />
Dr K. Hansen, Kryptogambotanik Naturhistoriska Riksmuseet, Box 50007, 104 05 Stockholm, Sweden; E-mail: karen.<br />
hansen@nrm.se<br />
Prof. dr K.D. Hyde, School of Science, Mae Fah Luang University, Tasud, Chiang Rai, Thailand; E-mail: kdhyde3@gmail.<br />
com<br />
Prof. dr L. Lange, Vice Dean, The Faculties of Engineering, Science and Medicine, Aalborg University; Director of Campus,<br />
Copenhagen Institute of Technology (CIT), Lautrupvang 15, DK-2750 Ballerup, Denmark; E-mail: lla@adm.aau.<br />
dk<br />
Prof. dr L. Manoch, Department of Plant Pathology, Faculty of Agriculture, Kasetsart University, Bangkok 10900, Thailand;<br />
E-mail: agrlkm@ku.ac.th<br />
Prof. dr W. Meyer, Molecular Mycology Research Laboratory, CIDM, ICPMR, Level 3, Room 3114A, Westmead Hospital,<br />
Darcy Road, Westmead, NSW, 2145, Australia; E-mail: w.meyer@usyd.edu.au<br />
Dr D. Minter, CABI Bioservices, Bakeham Lane, Egham, Surrey, TW20 9TY, UK; E-mail: d.minter@cabi.org<br />
Dr L. Norvell, Pacific Northwest Mycology Service, LLC, 6720 NW Skyline Boulevard, Portland, Oregon 97229-1309,<br />
USA; E-mail: llnorvell@pnw-ms.com<br />
Dr G. Okada, Microbe Division / Japan Collection of Microorganisms, RIKEN BioResource Center, 2-1 Hirosawa, Wako,<br />
Saitama 351-0198, Japan; E-mail: okada@jcm.riken.jp<br />
Prof. dr N. Read, Fungal Cell Biology Group, Institute of Cell and Molecular Biology, Rutherford Building, University of<br />
Edinburgh, Edinburgh EH9 3JH, UK; E-mail: nick@fungalcell.org<br />
Prof. dr K.A. Seifert, Research Scientist / Biodiversity (Mycology and Botany), Agriculture & Agri-Food Canada, K.W.<br />
Neatby Bldg, 960 Carling Avenue, Ottawa, ON, K1A OC6, Canada; E-mail: seifertk@agr.gc.ca<br />
Prof. dr J.W. Taylor, Department of Plant and Microbial Biology, University of California, 111 Koshland Hall, Berkeley,<br />
CA 94720, USA; E-mail: jtaylor@berkeley.edu<br />
Prof. dr M.J. Wingfield, Forestry and Agricultural Research Institute (FABI), University of Pretoria, Pretoria 0002, South<br />
Africa; E-mail: mike.wingfield@fabi.up.ac.za<br />
Prof. dr W.-Y. Zhuang, Systematic Mycology and Lichenology Laboratory, Institute of Microbiology, Chinese Academy of<br />
Sciences, Beijing 100080, China; E-mail: zhuangwy@sun.im.ac.cn<br />
<br />
i m a f U N G U S
ONE FUNGUS ONE NAME: A PLANT PATHOLOGIST’S<br />
VIEW<br />
Some eight months have gone by since the significant “One fungus = One name” (1F=1N) symposium was held at the<br />
historic home of the Netherland’s Academy of Science, Trippenhuis, in Amsterdam. This period might well go down in<br />
history as amongst the most traumatic, exciting, and important in the history of fungal taxonomy. The outcomes will undoubtedly<br />
influence the field and the many associated disciplines that rely upon it for decades, if not posterity.<br />
I<br />
had the privilege of being invited<br />
to present one of the introductory<br />
lectures at the 1F=1N symposium.<br />
Given the level of tension concerning the<br />
topic, I am not sure that I was particularly<br />
excited about doing so at the time. Yet as<br />
a plant pathologist and academic, I have<br />
had to grapple with the confusing <strong>issue</strong><br />
of fungi having more than one name,<br />
sometimes even different species names,<br />
for most of my career. Like many other<br />
plant pathologists, in my case working on<br />
trees, I have been confronted by farmers<br />
and foresters who have been frustrated,<br />
sometimes irritated, by the confusing<br />
names that they have had to contend with,<br />
while attempting to reduce the impact of<br />
plant diseases. Likewise, students in classes<br />
have been baffled by the complexities<br />
surrounding the dual nomenclature linked<br />
to anamorph/teleomorph connections and<br />
further complicated by pleomorphism in<br />
the asexual morphs of fungi. After many<br />
years of debate, often heated, the 1F=1N<br />
symposium in Amsterdam provided us with<br />
a perfect forum to debate the possibility of<br />
abandoning the dual nomenclature for the<br />
fungi.<br />
The turning point towards abandoning<br />
Article 59 and adopting a single name<br />
nomenclature arose as a result of DNA<br />
sequence data becoming increasingly<br />
available to mycologists. Effectively the<br />
“rules” dictated by the Code became<br />
restrictive, often redundant. Out of sheer<br />
frustration, some plant pathologists (see<br />
Crous et al. 2006) found it necessary to<br />
sidestep these regulations in order to present<br />
a meaningful taxonomy for important<br />
agents of plant disease. As John Taylor, the<br />
opening speaker at the 1F=1N symposium<br />
aptly stated, in seeking to abandon the dual<br />
nomenclature for fungi, we were in reality<br />
dealing with a situation where the proverbial<br />
“horse had already bolted” (Taylor 2011).<br />
My presentation followed Johns’ and, while<br />
also expounding on the inevitable demise of<br />
a dual nomenclature, as a plant pathologist<br />
and practitioner using mycology, my<br />
questions focussed mostly not on 1F=1N,<br />
but the <strong>issue</strong> of WHICH NAME we might<br />
most effectively use.<br />
The IF=1N symposium presented<br />
contradictory views regarding the<br />
opportunities and the hazards of<br />
abandoning a dual nomenclature for the<br />
fungi. The debates were lively, sometimes<br />
heated, and the event culminated in the<br />
drafting of the Amsterdam Declaration later<br />
published in this journal (Hawksworth et al.<br />
2011). It also precipitated the publication<br />
of a contrary view (Gams et al. 2011), which<br />
also had a large number of supporters.<br />
But most importantly, I believe that these<br />
debates provided the material that was<br />
essential for the all-important deliberations<br />
that would follow during discussions<br />
regarding the next edition of the Code at the<br />
International Botanical Congress that was to<br />
follow in Melbourne in July.<br />
The watershed discussions in Melbourne<br />
regarding the future of the taxonomy<br />
of the fungi are over. And the outcomes<br />
Mike Wingfield entering the rooms of the Royal Netherlands Academy of Arts and Sciences in Amsterdam for<br />
the One <strong>Fungus</strong> = One Name symposium on 19–20 April 2011.<br />
EDITORIAL<br />
volume 2 · no. 2<br />
(39)
EDITORIAL<br />
(Hawksworth 2011, Knapp et al. 2011,<br />
McNeill et al. 2011, Norvell 2011) are<br />
monumental to say the least. Some of us<br />
closely involved in the debate and that<br />
could not be in Melbourne received “blow<br />
by blow” accounts of the proceedings from<br />
Scott Redhead. Scott played an enormously<br />
important part in the debate, and had<br />
served as Secretary to a Special Committee<br />
established by the previous Congress in<br />
Vienna in 2005 to address this <strong>issue</strong> which<br />
had sadly failed to reach a consensus.<br />
From 1 January 2013, Article 59 will no<br />
longer provide for the separate naming of<br />
different morphs of the same fungus; all fungi<br />
will from then have only one correct name.<br />
I am absolutely convinced that that this will<br />
substantially promote our credibility with the<br />
practitioners of mycology. Certainly, in my<br />
situation, this will especially support plant<br />
pathologists and our stakeholders that rely on<br />
us to manage plant diseases (see Wingfield et<br />
al. 2011). But the real work post-Melbourne<br />
has yet to be done. The process towards<br />
deciding WHICH NAMES we use for the<br />
fungi now faces us, and another meeting at<br />
the Trippenhuis in April 2012 lies ahead to<br />
address this <strong>issue</strong>. Under the revised rules<br />
adopted in Melbourne, it will fortunately<br />
now be possible to develop approved lists<br />
of names with special protection to help<br />
minimize disruptions to the mycological<br />
community. Nevertheless, for some fungi, this<br />
is likely to be a road not entirely smooth. But<br />
it is a road towards a long-awaited natural<br />
classification for the fungi that had to be<br />
embarked on. Let the games begin.<br />
Crous PW, Slippers B, Wingfield MJ, Rheeder<br />
J, Marasas WFO, Philips AJL, Alves A,<br />
Burgess TI, Barber P, Groenewald JZ (2006)<br />
Phylogenetic lineages in the Botryosphaeriaceae.<br />
Studies in Mycology 55: 235–253.<br />
Gams W, Jaklitsch W, Agerer R, Aguirre-Hudson B,<br />
Andersen B, et al. (2011) A critical response to<br />
the ‘Amsterdam Declaration’. Mycotaxon 116:<br />
501–513.<br />
Hawksworth DL (2011) A new dawn for the<br />
naming of fungi: impacts of decisions made<br />
in Melbourne in July 2011 on the future<br />
publication and regulation of fungal names.<br />
MycoKeys 1: 7–20; <strong>IMA</strong> <strong>Fungus</strong> 2: 155–162.<br />
Hawksworth DL, Crous PW, Redhead SA, Reynolds<br />
DR, Samson RA, Seifert KA, Taylor JW,<br />
Wingfield MJ, et al. (2011) The Amsterdam<br />
Declaration on Fungal Nomenclature. <strong>IMA</strong><br />
<strong>Fungus</strong> 2: 105–112; Mycotaxon 116: 491–500.<br />
Knapp S, McNeil, J, Turland NJ (2011) Changes to<br />
publication requirements made at the XVIII<br />
International Botanical Congress in Melbourne<br />
— what does e-publication mean for you?<br />
Taxon 60: 1498–1501; Mycotaxon 117: in press.<br />
McNeill J, Turland NJ, Monro A, Lepschi B (2011)<br />
XVIII International Botanical Congress:<br />
preliminary mail vote and report of Congress<br />
action on nomenclature proposals. Taxon 60:<br />
1507–1520.<br />
Norvell LL (2011) Fungal nomenclature. 1.<br />
Melbourne approves a new Code. Mycotaxon<br />
116: 481–490.<br />
Taylor JE (2011) One <strong>Fungus</strong> = One Name: DNA<br />
and fungal nomenclature twenty years after<br />
PCR. <strong>IMA</strong> <strong>Fungus</strong> 2: 113–120.<br />
Wingfield MJ, de Beer ZW, Slippers B, Wingfield<br />
BD, Groenewald JZ, Lombard L, Crous<br />
PW (2011) One fungus one name promotes<br />
progressive plant pathology. Molecular Plant<br />
Pathology : in press.<br />
Michael J. Wingfield<br />
(Mike.Wingfield@fabi.up.ac.za)<br />
(40) ima fUNGUS
1000 fungal genomes to be sequenced<br />
fungi: 1000genomes<br />
As diverse decomposers, pathogens, and<br />
mutualistic symbionts, fungi have an<br />
enormous impact on human affairs and<br />
ecosystem function. Perhaps more than<br />
any other group of non-photosynthetic<br />
eukaryotes, fungi are essential biological<br />
components of the global carbon cycle.<br />
Collectively, they are capable of degrading<br />
almost any naturally occurring and<br />
manmade biopolymers. As such, fungi hold<br />
considerable promise in the development of<br />
alternative fuels, carbon sequestration and<br />
bioremediation of contaminated ecosystems.<br />
The use of fungi for the continued<br />
benefit of society requires an accurate<br />
understanding of how they interact in<br />
natural and synthetic communities. The<br />
ability to sample environments for complex<br />
fungal metagenomes is rapidly becoming<br />
a reality and will play an important part<br />
in harnessing fungi for industrial, energy,<br />
climate and ecosystem management<br />
purposes. Our ability to accurately analyze<br />
these data relies on well-characterized,<br />
foundational reference genome data of<br />
phylogenetically diverse fungi. With recent<br />
advancements in next generation sequencing<br />
technologies, the sequencing of fungal<br />
genomes is more tractable and less expensive<br />
than ever. The result is that sequencing of<br />
fungal genomes is becoming a relatively<br />
routine approach to data collection for all<br />
areas of mycology. This growth in genomics<br />
is resulting in a more integrated approach<br />
to biological research with basic tools of<br />
evolutionary biology (e.g., phylogenetic<br />
tree reconstruction, tests for selection, etc.)<br />
being central to comparative genomics.<br />
There exists a need, however, for systematic<br />
sampling of the Fungal Tree of Life to fully<br />
leverage our knowledge of fungal evolution<br />
and its application to genome-enabled<br />
mycology.<br />
To bridge this gap, an international<br />
research team in collaboration with the<br />
Joint Genome Institute ( JGI) of the US<br />
Department of Energy has embarked on a<br />
five-year project to sequence 1000 fungal<br />
genomes from across the Fungal Tree of<br />
Life. The team comprises Joseph Spatafora<br />
(Oregon State University), Jason Stajich<br />
(University of California at Riverside),<br />
Kevin McCluskey (Fungal Genetics Stock<br />
Center), Pedro Crous (KNAW-CBS Fungal<br />
Diversity Centre), Gillian Turgeon (Cornell<br />
University), Daniel Lindner (USDA Forest<br />
Service), Kerry O’Donnell and Todd Ward<br />
(USDA Agricultural Research Service),<br />
Antonis Rokas (Vanderbilt University), N.<br />
Louise Glass (University of California at<br />
Berkeley), A. Elizabeth Arnold (University<br />
of Arizona), Francis Martin (INRA,<br />
France), and Igor Grigoriev ( JGI). The<br />
overall plan is to fill gaps in the Fungal Tree<br />
of Life through the sequencing of at least<br />
two reference genomes from every accepted<br />
fungal family. One hundred species from<br />
27 orders and 55 families will be sequenced<br />
in the first year of sampling (Tier One)<br />
with ~225 additional species sequenced<br />
every year for the next four years. At the<br />
centre of this sampling effort are the unique<br />
biological resources that are housed in living<br />
culture collections, which provide access to<br />
high quality vouchered material.<br />
This is an exciting time as we move<br />
into the genomic era of mycology. This<br />
project has the core goal of providing<br />
reference information to inform<br />
research on comparative genomics,<br />
evolution of development, plant-microbe<br />
interactions, industrial mycology, and<br />
environmental metagenomic sequencing.<br />
Close cooperation with other large-scale<br />
genomic sequencing projects is underway<br />
and we envision building a network for<br />
global participation in the project. More<br />
information can be found on the project<br />
website ().<br />
Joey Spatafora<br />
(spatafoj@science.oregonstate.edu)<br />
NEWS<br />
The Ug99 stem rust (Puccinia graminis) of wheat<br />
threatens global supplies<br />
Ug99 is an aggressive race of<br />
stem rust of wheat (Puccinia<br />
graminis) first discovered in<br />
Uganda in 1998. The discovery<br />
was startling to pathologists<br />
and wheat breeders because<br />
the pathogen could potentially<br />
overcome the genetic resistance<br />
built into over half the world’s<br />
wheat crop. Ug99 is more<br />
dangerous than other rusts<br />
because of the number of<br />
resistance genes it is able to<br />
overcome.<br />
The world is starting to<br />
take notice of this global threat<br />
to food security. For example,<br />
even though Ug99 has not<br />
reached the USA, the threat is<br />
so urgent that the United States<br />
Department of Agriculture<br />
(USDA) recently surveyed<br />
Puccinia graminis race Ug99 on wheat. Photo Borlaug Global Rust Initiative.<br />
volume 2 · no. 2<br />
(41)
NEWS<br />
World distribution of Puccinia graminis race Ug99 on wheat (November 2011). Courtesy Borlaug Global Rust<br />
Initiative.<br />
2. Australia: <br />
• Oct-‐Nov 2010: Consistent air-flows<br />
from South Africa <br />
• Confirmed Ug99 (race PTKST: <br />
Sr31+Sr24 vir.) at source <br />
• Abnormal rainfall in Australia <br />
• SuscepMble hosts in Australia <br />
Emerging Concerns <br />
and identified varieties of wheat that show<br />
Ug99 resistance in efforts to prepare for the<br />
possibility of a Ug99 outbreak in the USA.<br />
The Durable Rust Resistance in Wheat<br />
(DRRW) project, funded by the Bill &<br />
Melinda Gates Foundation and the UK<br />
Department for International Development,<br />
and administered at Cornell University,<br />
has been established with the mission to<br />
mitigate this threat to the world’s wheat<br />
crop and avoid catastrophic losses. Two<br />
important DRRW objectives are pathogen<br />
surveillance and breeding wheat varieties<br />
that are resistant to the family of stem rust<br />
that includes Ug99, as well as stripe rust<br />
Jan-‐Mar 2011 <br />
Oct-‐Nov 2010 <br />
1. South Asia: <br />
• Jan-‐Mar 2011: Consistent air-flows<br />
from Yemen + Eritrea <br />
• Stem rust (Ug99?) at source in <br />
Yemen, Feb 2011 <br />
• High severity of stem rust <br />
Eritrea, Oct. 2010 <br />
• Highly suscepMble hosts in <br />
South Asia (PBW343: 6M ha; <br />
Inqualab-‐91: 4M ha) <br />
Possible spread of Puccinia graminis race Ug99 on wheat. Prepared by David Hodson (CIMMYT).<br />
(P. striiformis, syn. P. glumarum). New<br />
cultivated varieties must also improve<br />
farmers’ yields to keep pace with a growing<br />
world population.<br />
Ug99 is highly mobile, carried by wind<br />
or accidental human transmission. Because<br />
it does not recognize political boundaries,<br />
a surveillance system that is global in<br />
scope is necessary. The Global Cereal Rust<br />
Monitoring system is a coordinated network<br />
of national surveillance teams who collect<br />
standardized data from wheat fields in 20<br />
countries in Africa and Asia. The surveyors<br />
note any instance of rust, collect a sample<br />
of the rust for genetic testing, and record<br />
GPS data and other factors. These data are<br />
incorporated into a centralized database<br />
which allows researchers a bird’s eye view<br />
of the track of the pathogen, which helps in<br />
predicting where it might go next.<br />
Since its discovery in Uganda, Ug99<br />
has spread north-easterly through Kenya,<br />
Ethiopia, crossed the Red Sea into Yemen<br />
and Iran, and is now currently threatening<br />
Pakistan (where 24 M tons of wheat were<br />
produced in 2009) and India (over 80 M<br />
tons). Both countries have large plantings<br />
of wheat that are known to be susceptible.<br />
Historical evidence of the movements of<br />
rust pathogens indicates that Pakistan and<br />
India are at-risk. Ug99 has also traveled<br />
south, down the eastern coast of Africa to<br />
South Africa. There is a chance it could<br />
travel via wind currents over the Indian<br />
Ocean into Australia.<br />
Screening nurseries in Kenya and<br />
Ethiopia are key to the resistance breeding<br />
efforts. The nurseries are administered<br />
by CIMMYT (International Maize and<br />
Wheat Improvement Center), KARI<br />
(Kenya Agricultural Research Institute), and<br />
EIAR (Ethiopian Institute of Agricultural<br />
Research). Because Ug99 is present in east<br />
Africa, breeders from countries where Ug99<br />
is not present send promising new wheat<br />
varieties to these nurseries for field testing.<br />
Wheat breeders at the nurseries plant the<br />
seeds, expose them to Ug99, and collect<br />
screening data based on the incidence and<br />
severity of infection. If a variety is not<br />
resistant to Ug99 it soon becomes apparent.<br />
Thirty countries have sent germplasm for<br />
screening to KARI and EIAR, and 225<br />
000 lines of wheat have been screened since<br />
2006.<br />
Based on this screening process, 14<br />
varieties of wheat that are resistant to Ug99<br />
have been identified and are in testing with<br />
various national governments and seed<br />
programmes – and Ug99 resistant varieties<br />
have now been released in Ethiopia. Under<br />
an USAID (United States Agency for<br />
International Development) project, Ug99<br />
resistant seed has been distributed to six<br />
countries. A total of 12 000 ha of resistant<br />
varieties of wheat have been sown in Nepal,<br />
Bangladesh, Afghanistan, Egypt, Ethiopia,<br />
and Pakistan.<br />
Scientists on the DRRW project want to<br />
ensure that any new varieties released are not<br />
based on single gene resistance. Ug99, like<br />
any rust pathogen, has a great capability to<br />
(42) ima fUNGUS
change and evolve through mutation or sexual<br />
recombination. In fact, eight variants of Ug99<br />
have already been discovered. A wheat variety<br />
that only offers single gene resistance could be<br />
overwhelmed by Ug99 very quickly.<br />
The DRRW project also supports the<br />
Borlaug Global Rust Initiative (BGRI),<br />
founded by Norman Borlaug in 2005. The<br />
BGRI’s mission is to reduce the world’s<br />
vulnerability to rusts (stem, yellow, and leaf )<br />
and advocate for a sustainable international<br />
system for improving wheat yields and<br />
crop protection. The website for the BGRI<br />
() is regularly updated and<br />
should be consulted for more information.<br />
John Bakum<br />
BGRI Web Administrator and Content<br />
Developer<br />
( Jb755@cornell.edu)<br />
NEWS<br />
A DNA barcode for Fungi proposed for adoption<br />
At a meeting of the Fungal Working<br />
Group of the Consortium for the Barcode<br />
of Life (CBOL) held in Amsterdam on<br />
17–18 April 2011, it was agreed that a<br />
publication be prepared based on the<br />
studies carried out and which made a formal<br />
proposal to CBOL as to the barcode to<br />
be used for Fungi (see Schoch et al., <strong>IMA</strong><br />
Fungius 2: (5), June 2011). The results of<br />
an evaluation of six DNA regions by an<br />
international consortium using different<br />
groups of fungi, concluded that the internal<br />
transcribed spacer (ITS) region was the<br />
most appropriate as it had the highest<br />
probability of successful identification of<br />
the regions within the ribosomal cistron<br />
across the broadest range of Fungi. This<br />
was particularly valuable as the region to<br />
adopt as while it was not successful in all<br />
cases, it was the region that most commonly<br />
provided a barcode gap between the levels<br />
of within-species and between-species<br />
sequence variation. This region is therefore<br />
now to be formally proposed for adoption<br />
by the Consortium for the Barcode of<br />
Life as the primary fungal barcode marker,<br />
although it is recognized that supplementary<br />
barcodes may need to be proposed for<br />
adoption in some circumscribed taxonomic<br />
groups.<br />
The studies conducted involved the<br />
mitochondrial cytochrome c oxidase<br />
subunit 1, three regions from the nuclear<br />
ribosomal RNA cistron, regions of three<br />
representative protein coding genes (RPB1,<br />
RPB2 and MCM7), nuclear ribosomal small<br />
subunit (SSU), and nuclear ribosomal large<br />
subunit (LSU) – but all were inferior to<br />
the ITS. A multiauthored paper, involving<br />
148 researchers from 71 institutions, and<br />
describing the work carried out, is currently<br />
in revision in the Proceedings of the National<br />
Academy of Sciences of the USA, and is<br />
expected to appear shortly.<br />
New world record for the largest fungal basidiome<br />
A single basidiome of Rigidoporus ulmarius<br />
growing over the stump of an elm (Ulmus<br />
sp.) tree killed by Ophiostoma novo-ulmi<br />
in the grounds of the former International<br />
Mycological Institute at Kew (Surrey, UK)<br />
was accepted as the world’s largest in the<br />
Guiness Book of Records for 1990. That<br />
specimen measured 1.56 × 1.37 m and had<br />
a circumference of 4.53 m in January 1993<br />
(Aitchison & Hawksworth 1993). The<br />
basidiome continued to grow after that date,<br />
but it was never weighed, and subsequently<br />
deteriorated in situ.<br />
Now, an even larger single basidiome<br />
has been discovered on Hainan Island<br />
in southern China which is described<br />
by Dai & Cui (2011). This is of another<br />
polyporaceous fungus, Fomitiporia<br />
ellipsoidea, found on the underside of a<br />
fallen trunk of Cyclobalanopsis patellifornmis<br />
in virgin forest. This was judged to be 20 yr<br />
A portion of the basidiome with Yu-Cheng Dai.<br />
volume 2 · no. 2<br />
(43)
NEWS<br />
old and measured a massive 10.85 m long<br />
× 0.82–0.88 m wide and 4.6–5.5 cm thick,<br />
with a volume calculated as 409 000 – 525<br />
000 cm 3 and a fresh weight estimated at<br />
400–500 kg. With some 452 million pores,<br />
in an editorial note at the end of the article<br />
Nick P. Money suggests that it would have<br />
been generating around one trillion spores<br />
per day!<br />
Aitchison EM, Hawksworth DL (1993) IMI:<br />
retrospect and prospect. Wallingford: CAB<br />
International.<br />
Dai Y-C, Cui B-K (2011) Fomitiporia ellipsoidea<br />
has the largest fruiting body among the fungi.<br />
Fungal Biology 115: 813–814.<br />
A slice through a part of the basidiome showing<br />
the layers of pores.<br />
International meetings planned to discuss fungal<br />
<br />
nomenclature<br />
htt<br />
Following the dramatic revisions to<br />
nomenclatural rules for fungi enacted at<br />
the 2011 International Botanical Congress<br />
in Melbourne in July 2011 (Hawksworth<br />
2011, McNeill et al. 2011, Norvell 2011),<br />
members of both the Nomenclature<br />
Committee for Fungi (NCF) and the<br />
International Commission for the<br />
Taxonomy of Fungi (ICTF) have received<br />
many questions about how mycologists<br />
should adapt to the changes. Precise<br />
responses must wait publication of the new<br />
International Code of Nomenclature for<br />
algae, fungi, and plants which is expected<br />
in mid-2012, but the need for coordinated<br />
action is clear. With that in mind, the<br />
executives of these two bodies have agreed<br />
to cooperate to ensure the efficient sharing<br />
of accurate and consistent information and<br />
to develop inclusive, open, and transparent<br />
procedures for acting on the changes.<br />
Three major <strong>issue</strong>s are:<br />
(1) the migration away from<br />
dual nomenclature, based upon<br />
nomenclatural priority, community<br />
consensus and the best interests of users,<br />
via<br />
(2) the development of ‘Protected<br />
lists’ of accepted names and their<br />
nomenclatural types, together with<br />
those competing synonyms (including<br />
sanctioned names), and in some cases<br />
lists of rejected names, and<br />
(3) the establishment of permanent<br />
subcommissions or temporary working<br />
groups focused on specific narrowly or<br />
broadly delimited taxa to develop and<br />
recommend names for the ‘Protected’ or<br />
‘Rejected’ lists.<br />
Logically, some of these working groups will be<br />
derived from the permanent Subcommissions<br />
or associated Commissions of the ICTF<br />
(Aspergillus/Penicillium, Ceratocystis/<br />
Ophiostoma, Fusarium, Trichoderma/<br />
Hypocrea). Others are yet to be formed, and<br />
we hope that many will self-organize among<br />
existing taxonomic communities, and identify<br />
themselves to the NCF and ICTF, who can<br />
then provide support and information.<br />
As of this writing, four symposia are<br />
planned for 2012 to begin the process of<br />
sharing accurate information and formulate<br />
the expectations for the subcommissions<br />
and working groups:<br />
1F<br />
12–13<br />
=<br />
April<br />
2012<br />
?N<br />
CBS Symposium<br />
CBS symposium: One <strong>Fungus</strong> = Which<br />
Name? 12–13 April 2012, Trippenhuis,<br />
Royal Netherlands Academy of Arts<br />
and Sciences (KNAW), Amsterdam,<br />
the Netherlands. This meeting, which<br />
will focus entirely on understanding the<br />
nomenclatural changes and developing<br />
plans for implementation, will be a<br />
mixture of keynote speakers, breakout<br />
sessions, and book launches. It is unclear<br />
whether there will be any side meetings<br />
among the ICTF Subcommissions or<br />
other working groups associated with this<br />
symposium, although both the <strong>IMA</strong> and<br />
the ICTF plan to hold general meetings.<br />
Check the CBS website ()<br />
for more information.<br />
Mycological Society of Japan, 56th Annual<br />
Meeting. 26–27 May 2012, Gifu University,<br />
<br />
Gifu. This meeting will include a 2 hour<br />
symposium, “The new nomenclature - what<br />
is going on and what should be done.”<br />
Watch the MSJ website () for more information. A subsequent<br />
“Forum on microbial databases for academia<br />
and industry,” being organized by the<br />
Federation of Microbiology Societies, Japan<br />
(FEMS-Japan), is tentatively scheduled for<br />
29 May 2012 in Tokyo and will include<br />
presentations on several mycological<br />
databases.<br />
<br />
<br />
<br />
<br />
<br />
<br />
<br />
<br />
<br />
<br />
<br />
<br />
<br />
<br />
<br />
<br />
<br />
<br />
<br />
<br />
<br />
<br />
<br />
<br />
Mycological Society of America, Annual<br />
Meeting. 15–19 July 2012, Yale University,<br />
New Haven, CT, USA. This meeting will<br />
include a symposium, “The mycologist’s<br />
guide to the new International Code of<br />
Nomenclature for algae, fungi, and plants.”<br />
Preliminary plans are also underway<br />
for a satellite meeting of the ICTF<br />
Subcommission on Fusarium taxonomy<br />
(dgeiser@psu.edu) and a new Hypocreales<br />
working group to be held before or after<br />
the meeting (contact Amy Rossman,<br />
Amy.Rossman@ars.usda.gov). Watch<br />
<br />
the<br />
MSA website () for more<br />
information.<br />
<br />
<br />
<br />
<br />
<br />
<br />
<br />
<br />
<br />
<br />
<br />
<br />
<br />
<br />
<br />
<br />
(44) ima fUNGUS
INSTITUTE OF MICROBOLOGY CHINESE ACADEMY OF ...<br />
http://english.im.cas.cn/<br />
--><br />
Home | Sitemap | Contact | Directory | CAS | <br />
that all mycologists can indicate their Code Search: is the mandate of the NCF. The<br />
interest in specific taxonomic orders, in<br />
Please enter<br />
ICTF,<br />
keywords<br />
although focused primarily on<br />
About Us News People Research Education & Training International order Cooperation to facilitate Societies data & Publications sharing and Papers the Resources fungi Links of Join economic Us importance, has an<br />
Chinese Academy of Sciences, Institute of<br />
Microbiology Special Symposium: New<br />
development of working groups.<br />
Seminars<br />
The mandating of working groups,<br />
interest in facilitating the process for the<br />
broadest possible community. We welcome<br />
era of fungal nomenclature. Tentative dates<br />
9–10 August 2012, State Key Laboratory<br />
of Mycology, Beijing. Contact Dr. Cai Lei<br />
Genetic host resistance to PRRSV<br />
assurance of fair representation, avoidance participation from all mycologists.<br />
PresentorRaymond R. R. (Bob)<br />
of duplication of effort, and accommodation<br />
Rowland<br />
UniversityDiagnostic Medicine and<br />
of overlapping areas of interest are a priority. Pathobiology of Kansas Hawksworth State DL (2011) A new dawn for the<br />
University’s College of Veterinary<br />
(mrcailei@gmail.com), and consult the Working groups are encouraged to identify Medicine<br />
naming of fungi: impacts of decisions made<br />
Time09:30,11 23,2011<br />
June 2012 <strong>issue</strong> of <strong>IMA</strong> <strong>Fungus</strong> for more themselves and to form preliminary working Place:A323, Institute of Microbio... in Melbourne in July 2011 on the future<br />
information.<br />
relationships. Subcommissions of the ICTF<br />
Neural regulation of the immune<br />
publication and regulation of fungal names.<br />
system and its involvement in<br />
are expected to give these requirements top<br />
MycoKeys 1: 7-20; <strong>IMA</strong> <strong>Fungus</strong> 2: 155–162.<br />
inflammatory disease<br />
We encourage organizers of other<br />
PresentorLi Tian, PhD<br />
priority in their work plans, welcoming the McNeill J, Turland N, Monro AM, Lepschi<br />
mycological meetings to consider adding participation of other interested mycologists BJ (2011) XVIII International Botanical<br />
Papers<br />
News<br />
sessions Over-expression or symposia of an F-box protein on gene nomenclature CAI to Lei Honored <strong>IMA</strong> who Young might Mycologist not Award be [2011-11-10] formal members. Those<br />
Congress: preliminary mail vote and report of<br />
·Prof.<br />
reduces abiotic stress tolerance and<br />
Symposium on Advances Fungal Genomics and Evolution -- In Celebration of<br />
their · programs in 2012 in order to provide interested in forming such groups or in<br />
Congress action on nomenclature proposals.<br />
promotes root growth in rice ·Mini<br />
the Founding of the State Key Laboratory of Mycology [2011-10-26]<br />
Group II intron-anchored gene deletion in<br />
accurate information and encourage Host Species Of Famous sharing Tibetan concerns Medicinal <strong>Fungus</strong> should Identified communicate<br />
[2011-09-15]<br />
·<br />
Taxon 60: 1507–1520.<br />
Clostridium<br />
·Insect Multimedia<br />
Symposium Addresses Increased Demand for Industrial<br />
PARP and RIP-1 are required for autophagy<br />
participation by mycologists in all<br />
·TWAS-ROESEAP<br />
countries.<br />
Biotechnology [2011-08-29] with NCF Chair, Scott Redhead (scott. Norvell LL (2011) Melbourne approves a new Code.<br />
induced by 11’-deoxyverticillin A, which<br />
·<br />
precedes caspase-dependent apoptosis<br />
Executive Director Visits IMCAS [2011-07-22]<br />
·TWAS<br />
redhead@agr.gc.ca) and ICTF Chair, Keith Mycotaxon 116: 481–490.<br />
Probing genomic diversity and evolution of<br />
two Chinese ever Awarded the Bergey Medal [2011-05-25]<br />
·First<br />
The Streptococcus NCF suis and serotype ICTF 2 by NimbleGen are interacting Witnesses First Bergey’s Seifert Meeting (keith.seifert@agr.gc.ca).<br />
[2011-05-20]<br />
·<br />
tiling array<br />
·Beijing<br />
Launches World Data Center for Microorganisms [2011-05-18]<br />
regularly Functional evaluation with of the four putative curators of Index ·IMCAS<br />
Our goals are to ensure that all<br />
Scott Redhead (Chair) and<br />
DNA-binding regions in Thermoanaerobacter<br />
The 5th Intermational Day o...<br />
Fungorum ·<br />
tengcongensis reverse and gyrase MycoBank to ensure<br />
interested mycologists receive an accurate<br />
Lorelei Norvell (Secretary)<br />
Research Progress<br />
Chloropupukeanolides C–E, cytotoxic<br />
that pupukeanane the data chlorides required with a spiroketal for preparing Silencing in Regulation and of Interactions complete among understanding the Host, Virus Satellite<br />
·<br />
of RNA the new Societies & Publications Nomenclature Committee for Fungi (NCF)<br />
skeleton from Pestalotiopsis fici<br />
·RNA<br />
[2011-11-02]<br />
Protected Enhanced production lists of are recombinant available to working nomenclatural rules so that anyone wishing<br />
Comprehensive Structural Analysis of Neuraminidase Inhibition Using<br />
Nattokinase in Bacillus subtilis by promoter ·First<br />
groups · Laninamivir, a Highly Effective, Novel Influenza Drug [2011-10-26]<br />
and the community as a whole. to contribute may do so, and to define<br />
Keith Seifert (Chair) and<br />
SOD Achieve Industrialization [2011-10-17]<br />
·Hyperthermostable<br />
A nomenclatural database of connected of Non-cell-autonomous clearly Silencing the mechanisms of Endogenous Target and Gene expectations in for<br />
Andrew Miller (Secretary)<br />
·Restriction<br />
[More]<br />
Plants [2011-08-17]<br />
anamorph and teleomorph genera should participation. The format, standardization International Commission for the Taxonomy<br />
Genomic Analysis of Pathogenic and Evolution Mechanisms of<br />
·Comparative<br />
Streptococcus suis [2011-06-08]<br />
soon be placed on the <strong>IMA</strong> website<br />
and information requirements for the<br />
of Fungi (ICTF)<br />
Screening Strategy for Rapid Access to Polyether Antibiotics in<br />
·Genetic<br />
(). We intend Actinomycetes [2011-05-27] lists, and the approval of these final lists<br />
to develop a web-based mechanism so according to the requirements of the<br />
NEWS<br />
MycoKeys<br />
Institute Of Microbiology Chinese Academy of Sciences<br />
NO.1 West Beichen Road, Chaoyang District, Beijing 100101, China Phone: 0086-10-64807462 Fax: 0086-10-64807468 Email: office@im.ac.cn<br />
launched on 14 September 2011with the<br />
aim of accelerating biodiversity research,<br />
swelling further the options open for<br />
mycologists to publish their research. A sister<br />
journal to the already launched PhytoKeys<br />
and ZooKeys, MycoKeys will consider works<br />
describing new taxa, taxonomic revisions,<br />
checklists and catalogues, phylogenetic<br />
analyses, biogeography, methodology, data<br />
mining and literature surveys, monographs,<br />
atlases, letters and points of view, conference<br />
proceedings and Festschrifts, and data papers<br />
(manuscripts with large data sets). It is likely<br />
to be particularly attractive to authors of<br />
online to readers, though a few hard copies<br />
will be printed for archival purposes and<br />
deposited in key mycological libraries.<br />
Authors will, however, be able to purchase<br />
hard-copy offprints. There will normally<br />
be a fee of 15 € per page charge to authors,<br />
although this is being waived until September<br />
2012 on articles of up to 20 pages, and for<br />
the first 20 pages of longer works. Thorsten<br />
H. Lumbsch has been appointed Editor-in-<br />
Chief, and the publisher is Pensoft Publishers<br />
of Sofia, Bulgaria. Further information can<br />
be found on the journal’s website, .<br />
lengthy works or ones with many coloured<br />
1 of 1<br />
illustrations. The philosophy of the journal<br />
is discussed further in a leading article<br />
1<br />
Not to be confused with the very successful<br />
MycoKey 11/21/11 CD of keys 3:20 to the PMgenera of ascomycetes<br />
(Lumbsch et al. 2011), which includes a<br />
and basidiomycetes of Northern Europe produced<br />
The number of new mycological journals<br />
being produced continues to swell. MycoKeys 1<br />
is a new open-access mycological journal<br />
vision of automated connections of included<br />
data to key global databases and repositories.<br />
MycoKeys will be open-access and free<br />
by Thomas Læssøe & Jens H. Petersen (Version 2.1,<br />
2006; Version 3.1 Funga Nordica Edition, 2008); see<br />
.<br />
Lumbsch HT, Miller AN, Begerow D, Penev L (2011) MycoKeys, or why we need a new journal in mycology. MycoKeys 1: 1–6.<br />
volume 2 · no. 2<br />
(45)
REPORTS<br />
Genomics in China<br />
Mycologists at the Second Symposium for China’s Fungal Genome Initiative, Kunming. Photo by Juan Li.<br />
Mycologists at the Second Symposium for China’s Fungal Genome Initiative, Kunming. From left to right,<br />
Xingzhong Liu (Director of State Key Laboratory of Mycology at the Chinese Academy of Sciences’<br />
campus in Beijing and newly elected President of the Asian Mycological Association), John W. Taylor (<strong>IMA</strong><br />
President), Chungshu Wang (Chinese Institutes for Biological Sciences of the Chinese Academy of Sciences<br />
in Shanghai), Jiujiang Yu (Research Geneticist, USDA, SRRC in New Orleans), and Ke-Qin Zhang (Vice-<br />
President of Yunnan University). Photo taken with J. Taylor’s camera, photographer unknown.<br />
Mycologists at the Second Symposium for China’s Fungal Genome Initiative, Kunming. From left to right, Ke-<br />
Qin Zhang (Vice-President of Yunnan University), Joan Bennett (Rutgers University), Yunbo Qu (Shangdong<br />
University), and Zhiqiang An (University of Texas Health Science Center, Houston, Texas). Photo by Kaifang<br />
Ji.<br />
Two mycological symposia emphasizing<br />
fungal genomics were held in China<br />
in October 2011: a Mini-Symposium<br />
on Advances in Fungal Genomics and<br />
Evolution to celebrate the creation of the<br />
State Key Laboratory of Mycology at the<br />
Beijing campus of the Chinese Academy<br />
of Sciences; and the Second Symposium<br />
for China’s Fungal Genome Initiative,<br />
held at Yunnan University in Kunming.<br />
The meetings were organized by a trio of<br />
eminent Chinese mycologists: Xingzhong<br />
Liu (Director of the State Key Laboratory<br />
of Mycology in Beijing and newly elected<br />
President of the Asian Mycological<br />
Association), Chengshu Wang (Chinese<br />
Institutes for Biological Sciences of the<br />
Chinese Academy of Sciences in Shanghai<br />
and the vice President of the Mycological<br />
Society of China), and Keqin Zhang<br />
(Vice-President of Yunnan University).<br />
Many impressive presentations were made<br />
by Chinese scientists, which focused on<br />
fungi that parasitize insects and nematodes,<br />
including species of Cordyceps, Metarhizium,<br />
Beauveria, and Arthrobotrys. A visit by<br />
international symposium participants to the<br />
third largest freshwater lake in China, Taihu,<br />
brought home the importance of research<br />
into alternative methods of controlling<br />
agricultural pests in China – during an<br />
hour’s visit, only one, lone, immature gull<br />
was seen.<br />
The contingent of international<br />
visitors also visited the mycological<br />
facilities at the State Key Laboratory<br />
of Mycology, Institute of Microbiology<br />
in Beijing, and the Research Center<br />
for Insect Sciences, Institute of Plant<br />
Physiology and Ecology in Shanghai.<br />
In Beijing, more than 15 principal<br />
investigators conduct research in four<br />
areas: Diversity and Evolution; Functions<br />
and Interactions; Secondary Metabolism;<br />
and the Molecular Basis of Growth and<br />
Development. The four-story building<br />
that houses most of the PI’s labs also has a<br />
collection of more than 14 000 cultures, a<br />
herbarium with nearly 500 000 accessions,<br />
and a museum for fungi with interactive<br />
displays that hosts visits by groups<br />
of school children. In Shanghai, the<br />
collections and museum feature insects<br />
and the interaction of fungi and insects<br />
(46) ima fUNGUS
and the research facilities feature a stateof-the-art<br />
phytotron used for research on<br />
plant genetics and plant diseases caused<br />
by fungi.<br />
John W. Taylor<br />
President, International Mycological<br />
Association<br />
(jtaylor@berkeley.edu)<br />
International Association for Lichenology (IAL)<br />
REPORTS<br />
The IAL will hold its next quadrennial<br />
symposium in Bangkok on 9–13 January<br />
2012 () hosted by<br />
Ramkhamhaeng University. Most aspects<br />
of plant/microbial biology are represented<br />
amongst the session themes ranging<br />
from genomics and metabolites to<br />
forest ecology and global change.<br />
This is the first IAL conference to be<br />
hosted by a tropical nation, and reflects<br />
considerable interest and activity<br />
in lichen research by Thai scientists<br />
during the past 10 years. There are<br />
three post-symposium 5-day excursions<br />
and three workshops (Graphidaceae,<br />
Parmeliaceae, and Tropical lichens). Over<br />
300 abstracts for lectures and posters<br />
had been submitted by the submission<br />
deadline, promising an interesting and<br />
science-packed week. The Symposium<br />
is co-hosted by the universities of<br />
Chiang Mai, Mahasarakham, Maejo<br />
and Srinakarinwirot, The Biodiversity<br />
Research and Training Program, The Thai<br />
Botanical Society, The Thai Mycological<br />
Association, and The Queen Sirikit<br />
Botanical Garden.<br />
Peter D. Crittenden<br />
President, International Association for<br />
Lichenology<br />
(pdc@nottingham.ac.uk)<br />
Tourist hotel on stilts in a mangrove forest at Banpu where the post-conference Graphidaceae and Tropical<br />
Lichen workshops are to be held at IAL7.<br />
XVI Congress of European Mycologists (CEM XVI)<br />
The XVI Congress of European Mycologists<br />
was held in Porto Carras, Halkidiki,<br />
Greece, on 19–23 September 2011. This<br />
series of meetings is arguably the longest<br />
continuously running series of international<br />
congresses for mycology. Since its inception<br />
in Brussels in September 1956, it has visited<br />
widely different places within Europe, in<br />
accordance with a tradition of being hosted<br />
by a new country on each occasion. By<br />
coming to Greece in 2011, this was its first<br />
visit to this whole huge area of southeastern<br />
Europe, the Balkan Peninsula. It was also<br />
the first time ever on the shores of the<br />
Mediterranean. These congresses have<br />
always been arranged to ensure a balance is<br />
maintained between field and laboratory<br />
mycology. Very appropriately, therefore,<br />
the present congress, organized under<br />
the auspices of the European Mycological<br />
Association (EMA), and the sixteenth in the<br />
series, was at a resort surrounded by classic<br />
coastal aleppo pine woodland near the<br />
attractive seaside village of Neas Marmaras,<br />
about halfway down the eastern side of<br />
Sithonia, the central of the three long thin<br />
peninsulas which make Halkidiki such a<br />
distinct part of northern Greece.<br />
The Congress was presided over by<br />
the EMA President, and the Chair of the<br />
Organizing Committee was Stephanos<br />
Diamandis, the EMA Vice-President. The<br />
meeting was attended by 230 participants<br />
from 37 countries and every inhabited<br />
continent. After an ice-breaker party on<br />
the Sunday evening, formal sessions began<br />
on the Monday morning with a short<br />
opening ceremony and speeches of welcome<br />
from the local mayor, a representative of<br />
NAGREF, the main Congress sponsor<br />
in Greece, and the EMA President. The<br />
scientific programme comprised four days of<br />
lectures, presentations, workshops, symposia<br />
and posters, with one day, the Wednesday,<br />
reserved for field excursions, with a choice<br />
of two destinations. There was a plenary<br />
session each day, with keynote addresses,<br />
and these plenary sessions were followed<br />
each day by parallel sessions, poster sessions<br />
and satellite events covering a wide range of<br />
thematic areas. In addition to the scientific<br />
programme, there was, on the Thursday, a<br />
memorable Congress Dinner and, on the<br />
last day of the Congress, a business meeting<br />
of the General Assembly of the EMA.<br />
Plenary session keynote addresses<br />
• A new imaging nanotechnology for<br />
mycology (L. Kock).<br />
• Fungal conservation: insights from<br />
population biology and the impacts of<br />
past, present and future human land<br />
use (A. Dahlberg).<br />
• Fungal evolution: divergence and<br />
adaptation ( J. Taylor).<br />
• Fungal families: morphology,<br />
volume 2 · no. 2<br />
(47)
REPORTS<br />
Stephanos Diamandis making a presentation to <strong>IMA</strong><br />
President John W. Taylor.<br />
phylogeny and conflict resolution (P.F.<br />
Cannon).<br />
• MtDNA and rDNA: two different<br />
evolutionary lines combined for<br />
genetic differentiation, taxonomy<br />
and phylogeny in ascomycetes (M.A.<br />
Typas).<br />
• Outdoor airspora: patterns, prevalence<br />
and impacts (C. Rogers).<br />
Parallel sessions<br />
• Aeromycology (moderator E.<br />
Kapsanaki-Gotsi).<br />
• Alien and invasive fungi - biological<br />
control (moderator I. Kalucka).<br />
• Conservation of fungi (moderator C.<br />
Perini).<br />
• Developmental mycology (moderator<br />
R. Poeder).<br />
• Edible and medicinal fungi (moderator<br />
J. Baptista-Ferreira).<br />
• Fungal biotechnology (moderator J.<br />
Taylor).<br />
• Fungal distribution and diversity<br />
(moderators A. Abdel-Azeem, S.<br />
Diamandis, Z. Gonou-Zagou, and P.M.<br />
Kirk).<br />
• Fungal genetics and genomics<br />
(moderator M.A. Typas).<br />
• Fungi in ecosystems: effects of climate<br />
change (moderator L. Boddy).<br />
• <strong>Fungus</strong>-plant interactions: mycorrhizal<br />
systems (moderator R. Agerer).<br />
• Insect-fungus associations (moderator<br />
D. Schigel).<br />
• Plant pathogenic fungi (moderator J.<br />
Fatehi).<br />
• Systematics and evolution of fungi<br />
(moderators P.F. Cannon and C.<br />
Denchev).<br />
Poster sessions<br />
• Conservation of fungi, Developmental<br />
mycology, Systematics and evolution of<br />
fungi.<br />
• Aeromycology, Fungal distribution and<br />
diversity, Insect-fungus associations,<br />
Medical and veterinary mycology,<br />
Teaching mycology.<br />
• Biological control, Fungal<br />
biotechnology, Fungal genetics and<br />
genomics, Plant pathogenic fungi.<br />
Satellite events<br />
• Meeting of the European Council for<br />
Conservation of Fungi.<br />
• Symposium on application of IUCN<br />
criteria to fungi (moderator A.<br />
Dahlberg).<br />
• Workshop on conservation of<br />
ascomycetes (moderator D.W. Minter).<br />
Field excursions<br />
Wednesday 21 September, the middle<br />
day of the Congress, was devoted to two<br />
field excursions. The first was to Mount<br />
Holomon, in the main part of Halkidiki,<br />
some distance from the Congress, and<br />
at a higher altitude, which provided<br />
an opportunity to see fine broadleaved<br />
forests. The second, less than 10 km from<br />
the Congress site, was to the hill village<br />
of Parthenon from which an exploration<br />
of Mediterranean pine woodland, olive<br />
groves and scrub vegetation was planned.<br />
Both excursions were made but, very<br />
unfortunately, their start and finish<br />
coincided almost exactly with the passage of<br />
a front of torrential rain – the only rain in<br />
an otherwise warm and sunny week, making<br />
any serious field work practically impossible.<br />
This Congress was, scientifically, superb.<br />
Congresses of European Mycologists<br />
have always been strong in respect of<br />
biogeography, conservation, ecology, field<br />
mycology, recording and systematics.<br />
The world’s first group devoted to fungal<br />
conservation – the European Council for<br />
Fungal Conservation – arose from the<br />
1985 Congress of European Mycologists<br />
in Oslo and, in 2010, the EMA played a<br />
major role in establishing the International<br />
Society for Fungal Conservation, the only<br />
society anywhere exclusively devoted to<br />
protecting fungi. Not surprisingly, therefore,<br />
many of the sessions and satellite events<br />
at the Greek Congress demonstrated that<br />
the CEMs maintain a leading position in<br />
the now rapidly developing movement for<br />
fungal conservation. There has, however,<br />
been some concern within the EMA that<br />
other aspects of mycology have not always<br />
received sufficient attention at these events.<br />
At the preceding Congresses in Ukraine<br />
(2003) and Russia (2007) attempts were<br />
made, with increasing effectiveness,<br />
to address this imbalance. The Greek<br />
Congress not only continued that trend,<br />
but achieved a spectacular leap forward<br />
in terms of scientific coverage, with many<br />
aspects of laboratory-based mycology<br />
taking high-profile positions within<br />
the programme. The sessions on fungal<br />
biotechnology and on fungal genetics and<br />
genomics were highlights in that respect,<br />
but that did not mean that traditional<br />
themes were neglected. The session on<br />
insect-fungal interactions was memorable,<br />
and a particular achievement was to hold<br />
what appears to have been the first session<br />
devoted to aeromycology at a purely<br />
mycological congress.<br />
All in all, the Greek Congress has<br />
established a new high standard which<br />
will pose real challenges for subsequent<br />
Congresses to meet. To attract so many<br />
participants from so many different<br />
countries was a real achievement, not least<br />
because this was done at a time of severe<br />
recession and under the cloud of a huge<br />
global economic crisis. That achievement<br />
was a clear indication that the programme<br />
was as scientifically exciting as the location<br />
of the Congress was attractive. It was also<br />
reassuring to see many young mycologists<br />
present, and especially satisfying that,<br />
finally, a firm place has been established for<br />
Greek mycology not only on the European<br />
map, but also in the global arena. For this,<br />
mycology owes a great debt of gratitude to<br />
Stephanos Diamandis and his team on the<br />
Organizing Committee for their tireless<br />
work and huge generosity.<br />
David W. Minter<br />
President, European Mycological Association<br />
(d.minter@cabi.org)<br />
(48) ima fUNGUS
30 th ECCO Annual Meeting in Utrecht<br />
The European Culture Collections’<br />
Organisation (ECCO) held its 30 th Annual<br />
Meeting in Utrecht, The Netherlands,<br />
on 16–17 June 2011. The meeting was<br />
organized by the CBS curators and the<br />
ECCO board. With 71 participants from<br />
22 countries, it was the best attended annual<br />
meeting since the foundation of ECCO in<br />
1981. The ECCO Annual meetings bring<br />
together curators and other scientists from<br />
culture collections of all kinds throughout<br />
Europe to exchange the latest research,<br />
novel methods for strain identification and<br />
preservation, and to build networks and<br />
share ideas about the challenges faced by the<br />
collection community.<br />
The venue for this year’s meeting was<br />
Hotel Mitland, a choice much appreciated<br />
by the participants for the beautiful<br />
surroundings of the historical fortress<br />
‘De Bilt’ close to Utrecht city centre. The<br />
detailed programme of the meeting can be<br />
viewed on the ECCO website (), from where presentations can also<br />
be downloaded. On Thursday morning, the<br />
meeting was opened by CBS Director Pedro<br />
Crous and ECCO President Daina Eze,<br />
followed by four sessions, viz. phylogeny and<br />
taxonomy of microorganisms, developments<br />
in databases, and the Convention on<br />
Biological Diversity Nagoya protocol for<br />
Access and Benefit Sharing (ABS); the latter<br />
was followed by a round-table discussion<br />
on the consequences of ABS for microbial<br />
research. Possible strategies to influence the<br />
decisions national authorities will need to<br />
take that govern microbial exchange were<br />
also discussed. After the last session on<br />
collection network activities such as the<br />
EU-funded projects European Consortium<br />
Participants at the ECCO meeting in Utrecht.<br />
of Microbial Resources Centres (EMbaRC)<br />
and Microbial Resource Research<br />
Infrastructure (MIRRI), the party visited<br />
the CBS building on Thursday evening,<br />
where the conference dinner was also held.<br />
Joost Stalpers, who retired in May 2011<br />
after three decades of CBS curatorship,<br />
was addressed by former ECCO president<br />
Dagmar Fritze, thanking him on behalf of<br />
the entire ECCO community for his many<br />
contributions to the mission of ECCO.<br />
On Friday morning the meeting<br />
continued with a session presenting<br />
examples of successful collaborations<br />
between culture collections and industrial<br />
partners, followed by a series of talks on<br />
promising methods for strain identification<br />
and validation.<br />
After the official business of the<br />
ECCO General Meeting, poster prizes<br />
were awarded to the three best posters<br />
presented, viz. third prize to Celia Soares et<br />
al., second to Sashka Mihailova et al., and<br />
first to Marilia Maciel et al. Later that day,<br />
many participants joined in for a guided<br />
tour through Utrecht city centre, enjoying<br />
a selection of historic sites including the<br />
famous Dom tower. Many ECCO delegates<br />
also stayed on for a special meeting on<br />
Saturday 19 June organized by the Global<br />
Biological Research Centre Network<br />
demonstration project (GBRCN), where<br />
the preparations for MIRRI were discussed.<br />
The next ECCO meeting is to be<br />
held in Braga, Portugal, at the Micoteca<br />
Universidade do Minho (MUM), in June<br />
2012.<br />
Gerard Verkleij<br />
(g.verkleij@cbs.knaw.nl)<br />
REPORTS<br />
Mycological Society of America<br />
The Mycological Society of America held<br />
its 79 th annual meeting in the ‘Land of the<br />
Midnight Sun’ – Fairbanks, AK – during<br />
the first week of August 2011. Over 300<br />
mycologists were in attendance, many from<br />
foreign lands. Some attendees started the<br />
week with pre-meeting trips to Denali<br />
National Park or other local sites, enjoying<br />
the wilderness and big skies of Alaska.<br />
Others attended a two-day workshop,<br />
sponsored by the Fungal Environmental<br />
Sampling and Informatics Network<br />
(FESIN) on ‘Metamycology and beyond:<br />
using fungi in educational contexts’.<br />
The meeting kicked off with the<br />
Presidential Address by Thomas Bruns<br />
on ‘Revised thoughts on the processes<br />
that maintain local species diversity of<br />
ectomycorrhizal fungi.’ The invitational<br />
Karling Lecturer was Joseph Heitman<br />
from Duke University Medical Center.<br />
He gave a fascinating overview of the<br />
evolution of sex in fungi and spent<br />
the entire week interacting with MSA<br />
members. Symposia included: Mechanisms<br />
of fungal-plant interactions: perspectives<br />
from the interface of ecology, evolutionary<br />
biology, and genomics; Fungal population<br />
genetics; Diversification of fungi; Fungal<br />
contribution to organic matter storage<br />
volume 2 · no. 2<br />
(49)
REPORTS<br />
MSA Annual Foray, Fairbanks, participants. Photo by Andy Hart.<br />
and sequestration in soils; and Molecular<br />
ecology and biodiversity of arctic and boreal<br />
fungi. After spending the day listening to<br />
exciting contributed and invited talks, there<br />
were evening discussions on environmental<br />
sequencing and information from JGI<br />
on their fungal sequencing and analysis<br />
capabilities for individual researchers. The<br />
schedule was full, but there was still time<br />
to meet with colleagues for stimulating<br />
discussions of recent research.<br />
Several prominent mycologists received<br />
awards at the meeting. These included:<br />
Amy Rossman (USDA-ARS Systematic<br />
Mycology and Microbiology Laboratory,<br />
Beltsville, MD), Distinguished Mycologist;<br />
Lori Carris (Washington State University),<br />
Weston Teaching Award; Tim James<br />
(University of Michigan), Alexopoulus Prize<br />
for Outstanding Early-Career Mycologist;<br />
Greg Mueller (Chicago Botanical Garden),<br />
and Don Pfister (Harvard University),<br />
MSA Fellows; and Susumu Takamatsu<br />
(Mie University, Tsu, Japan), Honorary<br />
Member. Twenty-five students and young<br />
professionals received awards for travel and<br />
research excellence. The annual banquet<br />
included a wine-tasting competition, local<br />
cuisine, and entertainment by a Native<br />
American dance group. The annual auction<br />
featured a wide array of literature and<br />
mycological curiosities so that everyone left<br />
with something new (or in some cases, very<br />
old)!<br />
The foray was absolutely amazing<br />
with charismatic macrofungi everywhere!<br />
The morning was spent in the forests of<br />
the University of Alaska Large Animal<br />
Research Station among the musk ox and<br />
caribou. Later in the day, we left the campus<br />
to explore some of the local forests, which<br />
were also well stocked with mushrooms<br />
of many species. It was an amazing day<br />
and wonderful to visit with friends and<br />
colleagues among the trees.<br />
Preparations are already underway for<br />
next year’s annual meeting at Yale University<br />
in New Haven, Connecticut. The theme<br />
of that meeting will be ‘Integrative fungal<br />
biology: linking disciplines in basic and<br />
applied mycology.’ More information on<br />
this meeting can be found throughout 2012<br />
at . The east<br />
coast venue should provide easy access for<br />
our European colleagues, and we hope to see<br />
many of you there!<br />
Jessie A. Glaeser<br />
Secretary, Mycological Society of America<br />
(msasec1@yahoo.com)<br />
MSA Annual Foray, Fairbanks, display table. Photo<br />
by Andy Hart.<br />
D. Jean Lodge and friend (Boletus cf. edulis), MSA Annual Foray, Fairbanks. Photo by Jessie Glaeser.<br />
(50) ima fUNGUS
20 th Nordic Mycological Congress (NMC 20)<br />
The 20 th Nordic Mycological Congress<br />
(NMC) took place on the island Gotland<br />
in Eastern Sweden on 25 September to 1<br />
October 2011. It was organized by Ellen<br />
Larsson (University of Gothenburg), Mikael<br />
Jeppson, and myself (Swedish Museum of<br />
Natural History), and held in one of the<br />
Biological Sciences teaching laboratories<br />
at the University of Gotland in Visby. The<br />
NMC is primarily a congress for collecting<br />
and identifying fungi: mornings are used for<br />
collecting, and late afternoons and evenings<br />
for microscopic studies and discussions on<br />
identifications.<br />
Fifty-five professional and invited<br />
amateur mycologists attended, mainly from<br />
the Nordic countries (i.e. Denmark, Finland,<br />
Iceland, Norway, and Sweden) participated,<br />
but also including ones from China, Estonia,<br />
Scotland, and Spain joined. Gotland, situated<br />
in the Baltic Sea, is especially interesting<br />
because of its calcareous soil and warm<br />
climate. Many fungi are found here that are<br />
otherwise absent or rare in the rest of the<br />
Nordic countries. The meeting was a great<br />
success with plenty of fungi fruiting and<br />
beautiful sunshine all week. More than 32<br />
localities were visited, including a wide range<br />
of habitats, such as coniferous and deciduous<br />
forests, wooded meadows, alvars, and sand<br />
dunes. During the week 1525 collections<br />
were made, identified, and registered in a<br />
database (by Ibai Olariaga and PhD student<br />
Elisabeth Sjökvist, supported by the Swedish<br />
Taxonomy Initiative). The collections<br />
represented 596 species, including a large<br />
number Red Listed in Sweden. Additional<br />
collections identified within the three<br />
months after the meeting will be added and<br />
all information will be available in the Species<br />
Gateway database of the Swedish Species<br />
Information Centre ().<br />
Fungi were put out on display in the evening<br />
(in a tent placed outdoors to keep the fungi<br />
fairly fresh), and selected interesting groups<br />
of species were discussed. Several students<br />
from the universities of Gothenburg and<br />
Gotland attended the meeting and helped<br />
with the display. It was a very busy week, but<br />
more than anything the meeting provided an<br />
opportunity for mycologists to interact, enjoy<br />
and talk about fungi – and other topical<br />
<strong>issue</strong>s.<br />
The NMC is held every second year<br />
and rotates amongst the Nordic countries.<br />
Seppo Huhtinen (University of Turku) is<br />
the organizer of the next NMC, which is to<br />
be held in the Bear’s Lodge (Pohtimonlampi<br />
Hotel), close to Rovaniemi, Lappland, in<br />
northern Finland in 2013. Mycologists<br />
from outside the Nordic countries will be<br />
welcome.<br />
Karen Hansen<br />
(karen.hansen@nrm.se)<br />
REPORTS<br />
Scenes from the 20 th Nordic Mycological Congress on Gotland, Sweden, during field- and laboratory work, presentations, and one of the 47 Cortinarius subgen.<br />
Phlegmacium species collected, C. terpsichores. Photos by Tapio Kekki and Tor Erik Brandrud.<br />
volume 2 · no. 2<br />
(51)
AWARDS AND PERSONALIA<br />
AWARDS<br />
<strong>IMA</strong> Young Mycologist Awards 2011<br />
It is with great pleasure that the <strong>IMA</strong><br />
officers can now announce the winners of<br />
the first four of the inaugural International<br />
Mycological Association Young Mycologist<br />
Awards. The <strong>IMA</strong> Executive Committee<br />
had intended for then President Pedro<br />
Crous to present the awards last August<br />
at the XIV International Mycological<br />
Congress (IMC9) in Edinburgh. Alas, we<br />
learned that initiating the award process<br />
was far more complicated than we had<br />
foreseen. Now, with the process in place and<br />
functioning, we have received notification of<br />
the winners from Africa, Asia, Australasia,<br />
and North America. The committees<br />
for Europe and Latin America have been<br />
formed, and their winners will be reported<br />
in the first <strong>issue</strong> of <strong>IMA</strong> <strong>Fungus</strong> in 2012. All<br />
winners of the IMC9 round of <strong>IMA</strong> Young<br />
Mycologist Awards will receive their awards,<br />
which include a cheque for 500 €, at IMC10<br />
in Thailand in 2014.<br />
Please join me in congratulating the<br />
recipients: Marieka Gryzenhout (Ethel<br />
Mary Doidge Medal – African Regional<br />
Mycological Member Organization),<br />
Lei Cai (Keisuke Tubaki Medal –<br />
Asian Regional Mycological Member<br />
Organization), Ceri Pearce (Daniel<br />
McAlpine Medal – Australasian Regional<br />
Mycological Member Organization), and<br />
A. Elizabeth (‘Betsy’) Arnold (Arthur<br />
Henry Reginald Buller Medal – North<br />
American Regional Mycological Member<br />
Organization).<br />
John W. Taylor<br />
President, International Mycological<br />
Association (<strong>IMA</strong>)<br />
Ethel Mary Doidge Medal<br />
Marieka Gryzenhout, starting with<br />
her first paper published soon after she<br />
began graduate school, and continuing<br />
through to her book on an important<br />
plant pathogenic fungi (Gryzenhout et al.,<br />
2009, Taxonomy, Phylogeny, and Ecology of<br />
Bark-Inhabiting Tree-Pathogenic Fungi in<br />
the Cryphonectriaceae. St Paul, MN: APS<br />
Press). Marieka has contributed enormously<br />
to our knowledge of fungi. As a young<br />
mycologist with energy and intelligence, she<br />
has already made an impact in Africa as well<br />
as internationally. Her research has been<br />
particularly concerned with Diaporthales,<br />
an order which includes a number of serious<br />
forest pathogens, including chestnut blight<br />
fungus, Cryphonectria parasitica. She also<br />
discovered additional pathogens that threaten<br />
forests, and has determined the relationships<br />
between these species, describing new species<br />
and new genera where warranted, and also<br />
connecting sexual and asexual morphs.<br />
Marieka is also now investigating other plant<br />
pathogens, such as species of Phytophthora.<br />
She has assumed roles in several scientific<br />
societies, most spectacularly as the founding<br />
editor of the African Mycological Association<br />
(AMA) newsletter MycoAfrica, and is also<br />
now a member of the Executive Committee<br />
of the International Mycological Association<br />
(<strong>IMA</strong>). One of those supporting her<br />
nomination for this Medal commented that<br />
Marieka “represents the bright future for<br />
mycology in Africa.”<br />
Keisuke Tubaki Medal<br />
Lei Cai is an outstanding young mycologist<br />
in China. He has been working on the<br />
systematics and biodiversity of fungi from<br />
aquatic, coprophilous, and thermophilic<br />
habitats for many years. He has examined<br />
more than 5000 specimens, which has<br />
led to the discovery and descriptions of<br />
five new genera, and 45 new species. He<br />
is currently focusing on the systematics of<br />
plant pathogenic fungi using a polyphasic<br />
approach, and already has an impressive<br />
research record which includes one<br />
monograph, two book chapters, and 54<br />
peer-reviewed papers.<br />
For his excellent performance<br />
in mycological research, Lei Cai was<br />
awarded the prestigious Chinese Academy<br />
of Sciences (CAS) “Hundred-Talent<br />
Program”. He has also made significant<br />
contributions in promoting mycological<br />
studies in China and elsewhere in Asia. As<br />
Executive Associate Editor, he has played a<br />
key role in establishing and managing the<br />
new international journal, Mycology; he<br />
also serves as an associate editor of Fungal<br />
Diversity. He is an active educator in<br />
mycology who has given lectures at various<br />
international workshops or courses in the<br />
Chinese Academy of Sciences.<br />
Daniel McAlpine Medal<br />
Ceri Pearce was selected by the Australasian<br />
<strong>IMA</strong> Regional Mycological Organization for<br />
(52) ima fUNGUS
this Medal based on her research and major<br />
contributions to the mycological community.<br />
Her PhD research, which centred on the<br />
biology and taxonomy of Australian fungi,<br />
resulted in a book on Phyllachoraceae in<br />
Australia. More recently, with her research<br />
now focusing on the increasingly important<br />
area of biosecurity, she has been involved in<br />
the development of diagnostic and emergency<br />
response systems for exotic and introduced<br />
pests in Australia. Ceri has contributed to the<br />
mycologicsl community through her roles in<br />
mycology organizations, which includes the<br />
Australasian Mycological Society (executive<br />
councillor and librarian, 1998–2002),<br />
Australasian Plant Pathology Society<br />
(executive councillor 2005 –2007, and cochair<br />
of the Regional Councillor Working<br />
Group, 2005– 2008. In addition, she won<br />
the bid and successfully co-organised the 8 th<br />
International Mycological Congress (IMC8)<br />
in Cairns in 2006, which was the first time<br />
that an IMC had been held in the Southern<br />
Hemisphere. Finally, Ceri has been involved<br />
in numerous mycological educational and<br />
training programmes, both within her own<br />
organization and to agricultural industries,<br />
peers, and school students.<br />
Arthur Henry Reginald<br />
Buller Medal<br />
A. Elizabeth (‘Betsy’) Arnold received her<br />
PhD in Ecology and Evolutionary Biology<br />
in 2002 from the University of Arizona<br />
where she worked with Lucinda McDade.<br />
From 2003–2004, she was supported by<br />
a prestigious NSF Postdoctoral Fellow<br />
in Microbial Biology with sponsorship<br />
from François Lutzoni at Duke University.<br />
Betsy joined the faculty in the Division of<br />
Plant Pathology and Microbiology in the<br />
Department of Plant Sciences at the UA<br />
in 2005, where she is now an Associate<br />
Professor. Betsy currently teach courses<br />
in microbial diversity, plant sciences and<br />
mycology, curates the Robert L. Gilbertson<br />
Mycological Herbarium, and conducts<br />
research on the ecology, evolution and<br />
systematics of plant-associated fungi. She<br />
is best known for her innovative work in<br />
evolution and ecology of endophytes of<br />
terrestrial ecosystems. Her work is truly<br />
global in scale with sampling occurring<br />
along continental transects. It has resulted<br />
in our most complete understanding of the<br />
biodiversity and distribution of endophytic<br />
fungi and the World’s largest culture<br />
collection of endophytes. Betsy is a much<br />
sought after speaker across a spectrum of<br />
life science conferences, is an active member<br />
of the Mycological Society of America,<br />
and one of mycology’s biggest advocates.<br />
It is with great pride that North American<br />
mycologists acknowledge Betsy Arnold<br />
and her contribution to mycology with this<br />
Award.<br />
AWARDS AND PERSONALIA<br />
Distinguished Asian Mycologist Award<br />
Kevin D. Hyde was given the award of<br />
Distinguished Asian Mycologist in August<br />
2011 at the Asian Mycological Congress for<br />
his services in promoting Asian Mycology.<br />
Since 2008 Kevin has been Head and<br />
Associate Professor of the Institute of<br />
Kevin Hyde looking for freshwater fungi in southern France. Photo Jacques Fornier.<br />
Excellence in Fungal Research, School of<br />
Science, Mae Fah Luang University, Chiang<br />
Rai, Thailand and is also Managing Director<br />
of the Mushroom Research Foundation in<br />
Chiang Mai.<br />
He has held numerous prestigious<br />
positions in Asian mycological organizations,<br />
including EASIANET (of BioNet<br />
International) (Coordinator 2004–2007),<br />
Mycological Association of Hong Kong<br />
(Chair, 2002–2007), and <strong>IMA</strong> Asian<br />
Mycological Committee (Chair, 2007–<br />
2011). He founded and edited Fungal<br />
Diversity, a journal now pre-eminent in its<br />
field, and has been involved as an editor<br />
of about ten other journals. A prolific<br />
researcher, with over 600 publications and 15<br />
books to his credit, Kevin’s real passion is in<br />
training students – he has supervised some 20<br />
postdoctoral fellows, over 60 PhD students,<br />
and 15 MPhil students who have been drawn<br />
from Thailand, Sri Lanka, China, Laos,<br />
Myanmar, Vietnam, India, and Kenya.<br />
Kevin obtained his PhD from the<br />
University of Portsmouth, UK, under the<br />
volume 2 · no. 2<br />
(53)
AWARDS AND PERSONALIA<br />
Kevin Hyde teaching in the Mushroom Research Centre in Chiang Mai. Students from left to right: Marivic<br />
Cabanela, Nilam Wulandari, Iman Hidiyat, Subbu and unidentified.<br />
direction of E. B. Gareth Jones, in 1987.<br />
He first worked as a school teacher in<br />
Basingstoke, UK, and then Brunei, before<br />
moving to Australia where he surveyed<br />
plant pathogens in north Queensland<br />
and Papua New Guinea where he became<br />
fascinated by tropical microfungi. He<br />
subsequently obtained a tenured lectureship<br />
in the University of Hong Kong where<br />
he was based for 15 years before moving<br />
to Thailand. The Mushroom Research<br />
Foundation which he established also<br />
provides scholarships for PhD’s in mycology.<br />
Kevin strives to mould each of his students<br />
into renowned mycologists in their own<br />
right, and two of the first <strong>IMA</strong> Young<br />
Mycologists awards now made are to his<br />
previous students, Ceri Pearce and Lei Cai<br />
(see above).<br />
It is difficult to conceive of a more<br />
fitting recipient of this special award, and<br />
mycologists world-wide join in extending<br />
Kevin our congratulations and thanks for<br />
all he has done and continues to do for<br />
mycology. One of his current PhD students<br />
at Mae Fah Luang University, Samantha<br />
Karunarathna adds: “In Dr Hyde’s<br />
laboratory, we are budding mycologists, who<br />
have been taught, trained and mentored<br />
by Dr Hyde and would like to wish Dr<br />
Hyde all the very best and good luck in his<br />
endeavours to train and mould mycologists<br />
to salvage the world of mycology which is in<br />
dire need of many more mycologists.”<br />
BIRTHDAY GREETINGS<br />
Jiang-Chun Wei’s 80 th<br />
Jiang-Chun Wei celebrated his 80 th birthday<br />
on 6 November 2011. Born in Xianyang<br />
city, Shaanxi Province, China, his childhood<br />
to1945 was during World War II, and his<br />
middle-school life from 1945–1949 was the<br />
time of the Civil War of China. He majored<br />
in plant pathology at the Northwest<br />
Agricultural College from 1950, at a time<br />
when gene theory was opposed in China,<br />
but he never gave up his belief in science, for<br />
example, consulting the famous professor<br />
Sheng-Han Shi on the disease pathway<br />
of stripe rust of wheat from physiological<br />
and biochemical aspects. After graduating<br />
in 1955, Jiang-Chun was sent to work in<br />
the Institute of Agricultural Biology of<br />
Northwest China, of the Chinese Academy<br />
of Sciences (CAS). The first project for<br />
him was smut disease of millet, guided<br />
by the myco-pathologist Jian-Yi Li. Then<br />
in 1956 he was dispatched to the CAS<br />
Institute of Applied Mycology in Beijing<br />
where he studied the systematics of rust<br />
fungi on Rosaceae under the guidance of the<br />
mycologist Yun-Zhang<br />
Wang. In 1958, however,<br />
he was sent to Russia to<br />
finish his PhD degree<br />
under the guidance of the<br />
lichenologist Savicz at<br />
the Komarov Botanical<br />
Institute, in what was<br />
then Leningrad (now St<br />
Petersburg). He was soon<br />
publishing on lichens<br />
in Umbilicariaceae, a<br />
passion that stayed with<br />
him from that time,<br />
obtained his PhD in<br />
1962, and then returned<br />
to China. This time<br />
that was to the recently<br />
established CAS Institute of Microbiology<br />
in Beijing where he commenced work on<br />
the Chinese lichen biota in the Institute’s<br />
Laboratory of Mycology. His research<br />
career, however, was soon disrupted by an<br />
enforced absence from the Institute for<br />
ten years for the duration of the ‘Cultural<br />
Revolution’, after which he returned to<br />
the Institute of Microbiology, where he<br />
was Director of the Open Laboratory of<br />
Systematic Mycology and Lichenology<br />
(1985–1993) – the title of the laboratory<br />
(54) ima fUNGUS
eflecting his personal interests and which<br />
was one of the first in the world to embrace<br />
lichen-forming along with other fungi.<br />
Further, Jiang-Chun was the founding coeditor<br />
of Mycosystema (1986–1993). He<br />
also served as President of the Mycological<br />
Society of China (1993-2003), of which he<br />
was then made an Honorary President, and<br />
in 1995 he was awarded the degree of DSc<br />
by the Komarov Botanical Institute. To date,<br />
he has published 107 research papers, and<br />
also eight books, including An Enumeration<br />
of Lichens in China (1991, Beijing<br />
IN MEMORIAM<br />
International Science Publishers) and a<br />
monograph of his beloved Umbilicariaceae<br />
in Asia (with Jiang Yumei, 1993, The Asian<br />
Umbilicariaceae (Ascomycota), Beijing<br />
International Science Publishers).<br />
A celebration in his honour was held<br />
at the Institute of Microbiology on his<br />
birthday, where he gave a lecture ‘A glance<br />
back and prospect at my age of eighty’<br />
and enjoyed a huge celebratory cake.<br />
Jiang-Chung has been an inspiration to<br />
numerous students and fellow researchers<br />
in China, both for his vision, perseverance,<br />
and scientific contributions, and I feel<br />
honoured to have been privileged to know<br />
him and enjoy his hospitality in Beijing. All<br />
mycologists, including lichenologists, wish<br />
him a productive and fulfilling time.<br />
Xin Li Wei (Key Laboratory of Systematic<br />
Mycology and Lichenology, Institute<br />
of Microbiology, Chinese Academy of<br />
Sciences, Beijing) kindly provided much<br />
of the background information and<br />
photograph presented here.<br />
AWARDS AND PERSONALIA<br />
Aino Marjatta Henssen (1925–2011)<br />
A leading lichen taxonomist and systematist,<br />
Aino passed away on 29 August 2011. She was<br />
born on 12 April 1925 in Elberfeld, Germany,<br />
and had a German father and Finnish mother.<br />
She enroled at university to become a teacher,<br />
but due to her enthusiasm for research she<br />
graduated in 1953 with a doctoral thesis in<br />
plant physiology. After working in Bonn and<br />
Berlin at agricultural institutes investigating<br />
microorganisms decomposing manure, she<br />
started to work taxonomically – describing<br />
two new actinomycete genera and several<br />
new species. Following visits to Helsinki and<br />
Uppsala in 1956–61she turned her focus<br />
onto lichen fungi, inspired by the Swedish<br />
school of ascomycete systematics led by Johan<br />
Axel Nannfeldt, who had introduced ascoma<br />
development as a major character complex in<br />
ascomyete classification. Aino’s revision of the<br />
families Lichinaceae and Ephebaceae (1963,<br />
Symbolae Botanicae Upsaliensis 18<br />
(1): 1–123) is a prime example<br />
of this type of work, illustrated<br />
by numerous photographs of<br />
sections showing developmental<br />
stages. A decade later her textbook<br />
Lichenes: Eine Einführung in die<br />
Flechtenkunde (with her former<br />
PhD student Hans Martin Jahns,<br />
1973 1 , Stuttgart: Georg Thieme)<br />
she presented a classification<br />
that fully integrated lichenized<br />
with non-lichenized fungi, and<br />
introduced the “Zwischengruppe”<br />
for fungi that did not conform to<br />
the classical concepts of Nannfeldt;<br />
this was a remarkable work which<br />
presented ontogenetic data on all major groups<br />
based on her observations, which included<br />
many new findings, and descriptions of all<br />
families and orders she accepted<br />
In 1963 Aino was appointed curator of<br />
the herbarium at the Philipps-University in<br />
Marburg, and in 1970 as Associate Professor,<br />
a position she held until her retirement<br />
in 1990. Aino’s enthusiasm for lichens<br />
and other fungi attracted many students,<br />
and she continued her research as long as<br />
health permitted, with her last paper being<br />
published in 2007. In a series of more than<br />
120 publications, she described three orders<br />
(Arthoniales, Gyalectales, Lichinales), three<br />
families (Coccocarpiaceae, Coccotremataceae,<br />
Gloeoheppiaceae), 21 genera, and over<br />
150 new species, as well as numerous new<br />
combinations, often as a part of critical<br />
generic revisions. However, one of her mostcited<br />
works is her 1976 study with Peter W.<br />
James demonstrating by critical microscopic<br />
work that different morphologies could be<br />
produced by the same fungus depending on<br />
whether it has a cyanobacterial or a green–<br />
algal partner (in Brown DH et al. (ed.),<br />
Lichenology: progress and problems: 27–77,<br />
London: Academic Press). She was also the<br />
first researcher to appreciate the diversity of<br />
melanized non-lichenized rock-inhabiting<br />
fungi, describing many in Lichenothelia.<br />
Her 65 th birthday was marked with<br />
a ‘Festschrift’ ( Jahns HM (ed.), 1990,<br />
Bibliotheca Lichenologica 38: 1–427), and<br />
she was awarded the Acharius Medal of the<br />
International Association for Lichenology<br />
(IAL) in 1992. She will be remembered as<br />
a scientist who understood and pursued<br />
lichenology as a field of its own but also as<br />
an integral part of mycology.<br />
Prepared from a draft kindly provided by<br />
Heidi Döring (Royal Botanic Gardens,<br />
Kew, UK), and a PDF of a fuller obituary<br />
prepared along with H. T. Lumbsch which<br />
is to appear in The Lichenologist in 2012.<br />
1<br />
This work is dated “1974” but was actually<br />
published on 6 December 1973. Aino was always<br />
very concerned to point this out to show that her<br />
classification came out before that adopted by Josef<br />
Poelt (in Ahmadjian V, Hale ME (eds), 1974, The<br />
Lichens: 599–632, New York: Academic Press) –<br />
and which is dated “1973” but actually appeared on<br />
25 March 1974.<br />
volume 2 · no. 2<br />
(55)
AWARDS AND PERSONALIA<br />
Zang Mu (1930–2011)<br />
Zang Mu, one of the best-known Chinese<br />
macromycetologists, who was born on<br />
28 December 1930, passed away on 10<br />
November 2011, aged 81. A graduate<br />
from Soochow University, he worked at<br />
Nanjing Normal University as a teacher<br />
from 1954–1973, when he moved to the<br />
Kunming Institute of Botany (KIB) of<br />
the Chinese Academy of Sciences (CAS).<br />
As a research fellow in mycology and<br />
bryology, his major interests focused on<br />
systematics, ecology, and geography of<br />
fungi. He also studied mycorrhizas and their<br />
application in afforestation. He established<br />
the cryptogamic herbarium of KIB, and<br />
served as curator for many years. Other<br />
positions he was appointed to include that<br />
of Vice-President of the Mycological Society<br />
of China, and Vice-Director of the Key<br />
Laboratory of Mycology and Lichenology<br />
of CAS in Beijing. He published over<br />
150 papers on the fungi of China, and<br />
also several monographs, including the<br />
well-illustrated and remarkable [Economic<br />
Macrofungi from Southwestern China] (with<br />
Ying JZ, 1994, Beijing: Science Press), and<br />
had only this year completed [Dictionary<br />
of the Families and Genera of Chinese<br />
Cryptogams (Spore Plants)] (with Li XJ,<br />
2011, Beijing: Higher Education Press).<br />
Zang Mu twice received the secondclass<br />
Award in National Scientific and<br />
Technological Progress (1993, 1995), a<br />
second-class prize in China’s State Natural<br />
Science Award (2003), and also the N.<br />
Hiratsuka Award of the Mycological<br />
Society of Japan (2003). Besides mycology<br />
and bryology, he was also very interested<br />
in Chinese calligraphy, paintings, and<br />
collecting stamps. He contributed his full<br />
energy to the development of mycology and<br />
relative research fields in China, and is also<br />
warmly remembered for his kindness and<br />
hospitality to numerous visiting mycologists<br />
from overseas. China has lost a great<br />
mycologist, whose exceptional knowledge of<br />
Chinese mushrooms will be sorely missed.<br />
Prepared from material supplied by Liu<br />
Peigui and Zhu L. Yang (Kunming Institute<br />
of Botany, Chinese Academy of Sciences,<br />
Kunming, China).<br />
(56) ima fUNGUS
Horizontal Gene Transfer (HGT) from Fungi is the<br />
basis for plant pathogenicity in oomycetes<br />
It always seemed rather odd that some<br />
oomycetes (Oomycota) were so fungal-like<br />
in their behaviour as plant pathogens.<br />
Now whole-genome comparisons are<br />
starting to reveal just why. Richards et al.<br />
(2011) have undertaken a painstaking<br />
gene-by-gene analysis of the proteomes<br />
of Hyaloperonospora parasitica and three<br />
species of Phytophthora (P. infestans, P.<br />
ramorum, and P. sojae) which reveals<br />
an extensive pattern of cross-kingdom<br />
horizontal gene transfer (HGT) from Fungi.<br />
In the case of P. ramorum, an amazing 7.6<br />
% of the secreted proteome appears to have<br />
been acquired from true fungi. In all, 34<br />
cases of HGT were identified, of which the<br />
case for 21 was strongly supported, most<br />
of which seem to have occurred close to<br />
the shift from phagotrophy to osmotrophy<br />
and the evolution of the fungal cell wall at<br />
the base of the monophyletic Fungi clade.<br />
Amongst the genes evidently transferred,<br />
are one related to features such as the ability<br />
to break down plant cell walls and take<br />
up sugars, nitrogen and phosphates, and<br />
further ones implicated in overcoming plant<br />
defence mechanisms and attacking plant<br />
cells. A schematic diagram of the functional<br />
proteome of oomycetes derived from<br />
Fungi is provided (their Fig. 2) at which<br />
one can only marvel at the complexity. The<br />
phenomenon was already recognized some<br />
years ago by Richards et al. (2006), but only<br />
five HGT gene transfers were reported at<br />
that time. This more detailed study, made<br />
possible by increasingly available genome<br />
sequences, reveals that this phenomenon<br />
is much more extensive than might have<br />
been imagined. Thus, it is not a matter of<br />
oomycetes merely being ‘fungal analogues’<br />
or ‘pseudofungi’, they are actually partly<br />
Fungi.<br />
It should be noted that HGT is not<br />
only unidirectional from Fungi into to<br />
other organisms. Indeed, in another paper<br />
published this summer, this same group of<br />
researchers report identifying 323 examples<br />
of HGT into Fungi from prokaryotes<br />
(principally bacteria) and also other Fungi<br />
(Richards et al. 2011b).<br />
RESEARCH NEWS<br />
Richards TA, Dacks JB, Jenkinson JM, Thornton CR, Talbot NJ (2006) Evolution of filamentous plant pathogens: gene exchange across eukaryotic kingdoms. Current<br />
Biology 16: 1857–1864.<br />
Richards TA, Leonard G, Soanes DM, Talbot NJ (2011b) Gene transfer into fungi. Fungal Biology Reviews 25: 98–110.<br />
Richards TA, Soanes DM, Jones MDM, Vasieva O, Leonard G, Paszkiewicz K, Foster PG, Hall N, Talbot NJ (2011a) Horizontal gene transfer facilitated the evolution<br />
of plant parasitic mechanisms in the oomycetes. Proceedings of the National Academy of Sciences, USA 108: 15258–15263.<br />
1 HGTs<br />
8 HGTs<br />
2 HGTs<br />
1 HGTs<br />
9 HGTs<br />
Saccharomycotina<br />
Pezizomycotina Fungi<br />
‘other fungi’<br />
Metazoa<br />
Amoebozoa<br />
Archaeplastida<br />
Alveolata<br />
Blastocystis hominis<br />
Diatoms<br />
Ectocarpus siliculosus<br />
Aureococcus anophagefferens<br />
Hyphochytrium catenoides<br />
Saprolegnia parasitica<br />
Albugo laibachii<br />
Phytophthora ramorum<br />
Phytophthora sojae<br />
Phytophthora infestans<br />
Hyaloperonospora parastica<br />
Unikonts<br />
S<br />
t<br />
r<br />
a<br />
m<br />
e<br />
n<br />
o<br />
p<br />
i<br />
l<br />
e<br />
s<br />
Schematic figure showing the pattern of horizontal gene transfer (HGT) between fungi and oomycetes, adapted from Richards et al. (2011a: fig. 1). In total Richards et<br />
al. (2011) provide evidence of 34 gene transfer events. Using phylogenetic methods combined with alternative topology tests they polarised the ancestry of 21 transfer<br />
events and are illustrated in this figure. The figure shows the transfers were generally concordant with the diversification of plant parasitic oomycetes, consistent with<br />
putative annotation of many of these genes which suggest that they are important for plant parasitism. Figure courtesy of Tom A. Richards.<br />
volume 2 · no. 2<br />
(57)
RESEARCH News<br />
Fungal pathogens as a driver of tree species<br />
diversity in tropical forests<br />
It has been postulated that at least one of<br />
the factors promoting the maintenance of<br />
diversity in tropical forests is the action<br />
of host-specific parasites and pathogens<br />
which are more likely to kill seedlings<br />
near the parent tree species, the so-called<br />
Janzen-Connell ( J-C) model. In order to<br />
test whether this held for plant pathogens,<br />
Konno et al. (2011) isolated Colletotrichum<br />
anthrisci from seedlings of four trees<br />
growing below the same tree species<br />
where they had been killed by dampingoff<br />
beneath: Cornus controversa, Fraxinus<br />
lanuginosa, Magnolia obovata, and Prunus<br />
grayana. The isolates from all four species<br />
were confirmed as identical by 99–100 %<br />
similarity in ITS sequences (5.8SrDNA,<br />
ITS1 and ITS2), and inoculated into<br />
seedlings of F. lanuginosa and Prunus<br />
grayana. In all cases some damage to the<br />
seedlings occurred, but this was most severe<br />
in seedlings inoculated with isolates from<br />
the conspecific host tree. This suggests a<br />
degree of specialization whereby seedlings<br />
from the same tree species are more<br />
likely to be eliminated than those from<br />
different hosts – consequently reducing<br />
the probability of seedlings of the same<br />
tree species surviving when growing near<br />
examples of the same species. Konno et al.<br />
postulate that if this situation is common<br />
within several pathogens in a mixed<br />
tree forest, then diversity will tend to be<br />
maintained as proposed in the J-C model.<br />
The study also shows that fungi with<br />
identical ITS sequences obtained from<br />
native forest trees can differ in their degree<br />
of pathogenicity to other tree species.<br />
Konno M, Iwamoto S, Seiwa K (2011) Specialization of a fungal pathogen on host tree species in a cross-inoculation experiment. Journal of Ecology 99: 1394–1401.<br />
Colletotrichum anthrisci (from ex-type strain CBS 125334). a–b. acervuli; c. tip of a seta; d–e. conidiophores; f. conidiophores and setae; g–i. appressoria; j–k.conidia;<br />
all from ex-type culture CBS 125334. a, c–e, j: from Anthriscus stem; b,f, g, k: from SNA. a–b: DM; c–k: DIC. — Scale bars: a = 200 μm; e = 10 μm; a applies to a–b; e<br />
applies to c–k. Photos courtesy Ulrike Damm.<br />
(58) ima fUNGUS
A new system for the arbuscular mycorrhizal fungi<br />
(Glomeromycota)<br />
In the course of the last ten years, and<br />
especially during the last five, immense<br />
progress has been made in understanding<br />
the molecular phylogenetic relationships<br />
of arbuscular mycorrhizal fungi, members<br />
of the phylum Glomeromycota. As might<br />
have been expected for a group which was<br />
already represented by modern-looking<br />
representatives in the Devonian, and which<br />
forms mutualistic associations with some<br />
80--85 % of land plants around today, there<br />
was much diversity to be detected. In 2011<br />
a succession of key papers describing new<br />
classes, families, and genera has appeared,<br />
mainly prepared by Fritz Oehl (Zürich,<br />
Switzerland), Gladstone Alves da Silva<br />
(Recife, Brazil), and Javier Palenzuela<br />
(Granada, Spain), with various colleagues,<br />
has appeared (e.g. Oehl et al. 2011ad).<br />
Building on the pioneering work of<br />
Christopher Walker, Arthur Schüβler,<br />
and James B. Morton in particular, these<br />
researchers have established robust<br />
correlations between microscopic features<br />
and the major groupings emerging from<br />
molecular studies. An elegant consumerfriendly<br />
digest and synthesis of the new<br />
system for the phylum has now been<br />
prepared, which we are proud to include in<br />
the current <strong>issue</strong> of <strong>IMA</strong> <strong>Fungus</strong> (Oehl et<br />
al. 2011e).<br />
In the new system, three classes<br />
(Archaeosporomycetes, Glomeromycetes,<br />
and Paraglomeromycetes), five orders<br />
(Archaeosporales, Diversisporales,<br />
Gigasporales, Glomerales, and<br />
Paraglomerales), 14 families, and 29<br />
genera are recognized. Key anatomical<br />
and morphological features characterizing<br />
the molecularly supported taxa are spore<br />
formation, the number of spore walls,<br />
germination type and structure, and<br />
mycorrhizal structures (stained in Trypan<br />
blue). These characters are illustrated<br />
and tabulated down to genus level in the<br />
synthesis paper, and using this many genera<br />
will now be separable using light microscopy<br />
alone. This paper is set to become the<br />
key reference work on this remarkable<br />
fungal phylum for ecologists and others<br />
investigating or utilizing endomycorrhizal<br />
fungi.<br />
RESEARCH NEWS<br />
Oehl F, Silva GA, Goto BT, Sieverding E (2011a) Glomeromycetes: three new genera and glomoid species<br />
reorganized. Mycotaxon 116: 75–120.<br />
Oehl F, Silva DKA, Maia LC, Sousa NMF de, Vieira HEE, Silva GA (2011b) Orbispora gen. nov., ancestral in<br />
the Scutellosporaceae (Glomeromycetes). Mycotaxon 116: 161–169.<br />
Oehl F, Silva GA, Goto BT, Maia LC, Sieverding E (2011c) Glomeromycota: two new classes and a new order.<br />
Mycotaxon 116: 365–379.<br />
Oehl F, Silva GA, Sánchez-Castro I, Goto BT, Maia LC, Vieira HEE, Barea JM, Sieverding E, Palenzuela J<br />
(2011d) Revision of Glomeromycetes with entrophosporoid and glomoid spore formation with three new<br />
genera. Mycotaxon 117: 297–316.<br />
Oehl F, Sieverding E, Palenzuela J, Ineichen K, Silva GA (2011e) Advances in Glomeroycota taxonomy and<br />
classification. <strong>IMA</strong> <strong>Fungus</strong> 2:191—199.<br />
Examples of characteristics of spore bases and subtending hyphae in Glomeromycota. A. Glomus ambisporum.<br />
B. G. aureum. C. Funneliformis coronatus. D. Septoglomus constrictum. sw = spore wall layers; sp = bridging<br />
septum; sh = subtending hypha. See Oehl et al. (<strong>IMA</strong> <strong>Fungus</strong> 2: 191–199, 2011) for further explanation.<br />
Photos courtesy Fritz Oehl.<br />
New insights into global fungal species numbers?<br />
Blackwell (2011) has revisited the <strong>issue</strong> of<br />
how many fungi exist on Earth, and the<br />
impact that molecular studies, and especially<br />
high-throughput environmental sequencing<br />
has had on our understanding of the extent<br />
of that diversity. She draws attention to the<br />
state of knowledge of the fungi in particular<br />
habitats, and the <strong>issue</strong> of phylogenetic<br />
species not or hardly separable by other<br />
features. Attention is drawn to the increased<br />
number of flowering plants suggested<br />
to exist beyond the 270 000 used in the<br />
extrapolations of Hawksworth (1991): for<br />
example, Paton et al. (2008) provide a figure<br />
of 352 000 for known species, and Joppa<br />
volume 2 · no. 2<br />
(59)
RESEARCH News<br />
1.5 M + ?<br />
et al. (2010) expect that to grow by 10–20<br />
% – i.e. to 387 200–422 400 species. If these<br />
larger figures are used, then the 6 : 1 ratio of<br />
fungi : plants used by Hawksworth (1991)<br />
would yield a figure of 2.3–2.5 M species of<br />
fungi.<br />
Blackwell also emphasises the results<br />
of the study of O’Brien et al. (2005) from<br />
environmental sequences from soil which<br />
suggested a total number of 3.5–5.1 M<br />
fungus species. However, results from highthroughput<br />
sequencing studies have to be<br />
approached with some caution as critical<br />
studies with bacterial populations suggest<br />
that they may overestimate the number of<br />
taxa actually present by a factor of about six<br />
(Quince et al. 2009). If it were justified to<br />
apply that factor in the O’Brien et al. study,<br />
their figure would not have been so far<br />
ahead of other estimates.<br />
A different approach to estimating<br />
global species numbers of all organisms was<br />
taken by Mora et al. (2011), who found<br />
that the description of taxa according to<br />
rank followed a predictable pattern across<br />
different groups of organisms. In the case of<br />
the fungi, the validity of the approach might<br />
be questioned as our knowledge is so poor,<br />
and also as they worked with a figure of just<br />
43 271 species of fungi – the number in the<br />
Catalogue of Life 2010 Annual Checklist<br />
(Bisby et al. 2010) rather than the 100 000<br />
figure currently in general use (e.g. Kirk et<br />
al. 2008). However, what was intriguing is<br />
that based on that data set, they predicted<br />
611 000 (± 297 000) fungal species which<br />
implies that only around 7 % (range<br />
4.5–13.5 %) are now known, perhaps not so<br />
different from the 5 % previously proposed<br />
(Hawksworth 1991).<br />
While it now seems that the 1.5 M<br />
estimate may indeed be conservative, as<br />
stated when it was proposed (Hawksworth<br />
1991), the jury remains out as to how much<br />
by. For that reason I am inclined to still<br />
work with “at least 1.5 M” as the additional<br />
factor is so uncertain, as I concluded a<br />
decade ago (Hawksworth 2001). Much<br />
more field-truthed data are needed to<br />
improve the current estimates, and as<br />
Blackwell points out, this “can be speeded<br />
by enlisting more biologists to accomplish<br />
the goal” (p. 434). But how can that be<br />
done? The answer may lie in an increasing<br />
recognition within the scientific community<br />
of the scale of the problem, the excitement<br />
of discovering novel taxa, and a hightened<br />
appreciation of the crucial role of fungi in<br />
so many aspects of human concern from<br />
health to food security and climate change.<br />
A paradigm shift in the foci of biological<br />
research may be required.<br />
Bisby FA, Ropskov YR, Orrell TM, Nicolson D, Paglinawan LE, et al. (2010) Species 2000 and ITIS Catalogue of Life: 2010 annual checklist. Reading: Species 2000.<br />
Blackwell M (2011) The Fungi: 1, 2, 3 . . . . 5.1 million species. American Journal of Botany 98: 426–438.<br />
Hawksworth DL (1991) The fungal dimension of biodiversity: magnitude, significance, and conservation. Mycological Research 95: 641–655.<br />
Hawksworth DL (2001) The magnitude of fungal diversity: the 1.5 million species estimate revisited. Mycological Research 105: 1422–1432.<br />
Joppa LN, Roberts DL, Pimm SL (2010) How many species of flowering plants are there? Proceedings of the Royal Society of London, B, 278: 554–559.<br />
Kirk PM, Cannon PF, Minter DW, Stalpers JA (2008) Ainsworth & Bisby’s Dictionary of the Fungi. 10 th edn. Wallingford: CAB International.<br />
Mora C, Tittensor DP, Adl S, Simpson AGB, Worm B (2011) How many species are there on Earth and in the Ocean? PLoS Biology 9(8): e1001127.<br />
O’Brien BL, Parrent JL, Jackson JA, Moncalvo JM, Vilgalys R (2005) Fungal community analysis by large-scale sequencing of environmental samples. Applied and<br />
Environmental Microbiology 71: 5544–5550.<br />
Paton AJ, Brummitt N, Govaerts R, Harman K, Hinchcliffe S, Allkin B, Lughadha EM (2008) Towards Target 1 of the Global Strategy for Plant Conservation: a<br />
working list of all known plant species – progress and prospects. Taxon 57: 602–611.<br />
Quince C, Lanzén A, Curtis TP, Davenport RJ, Hall N, Head IM, Read IF, Sloan WT (2009) accurate determination of microbial diversity from 454 pyrosequencing<br />
data. Nature Methods 6: 639–641.<br />
Powdery mildews under scrutiny<br />
The Special Interest Group (SIG) meetings<br />
held during IMC9 in August 2010 were<br />
exceedingly popular. Mini-reviews based on<br />
the SIG events have already been published<br />
in previous <strong>issue</strong>s of <strong>IMA</strong> FUNGUS, and<br />
a longer review on molecular diagnostic<br />
methods is included in this <strong>issue</strong> (pp.<br />
177–189). In the case of the SIG on<br />
powdery mildew fungi (Erysiphales),<br />
convened by Uwe Braun, Levente Kiss and<br />
Susumu Takamatsu, the papers presented<br />
are published as an <strong>issue</strong> of Mycoscience 52<br />
(3) (May 2011) under the title ‘Biology,<br />
biodiversity, evolution and systematics of<br />
the Erysiphales’.<br />
The focus on the papers is on the<br />
biology and evolution of pathogenesis and<br />
variation at the molecular level in particular<br />
species or complexes. Aleš Lebeda et al. (pp.<br />
59—164) review the pathotype and race<br />
designations and tests used in two powdery<br />
mildews attacking cucurbits, Golvinomyces<br />
cichoracearum and Podosphaera xanthii.<br />
The situation has become confusing as<br />
a result of the use of different Cucumis<br />
species and cultivars in challenge testing,<br />
with, for example, three systems being<br />
used by different research groups for race<br />
denomination; the adoption of two sets of<br />
host genotypes and a concise designation<br />
system are commended. Quercus is host<br />
to more powdery mildews than any other<br />
genus, with over 50 species recognized on<br />
members of the genus. Those present on oak<br />
in Europe, which had all been considered<br />
as introduced, are reviewed by Marie-Laure<br />
Desprez-Loustau et al. (pp. 165–173).<br />
Molecular phylogenetic studies confirm<br />
that within Eryisphe three species are<br />
(60) ima fUNGUS
involved; E. alphitoides (syn. Microsphaera<br />
alphitoides) which is the most pathogenic,<br />
E. hypophylla, and E. quercicola recently<br />
discovered in France and only known<br />
from a single ITS sequence. Affinities with<br />
powdery mildews known from tropical<br />
hosts discovered were a surprise, and hostjumping<br />
and specialization on Q. robur is<br />
postulated to have occurred in the evolution<br />
of E. alphitoides. Molecular phylogenetic<br />
studies on Erysiphe species on Ligustrum<br />
and Syringia (both Oleaceae) by Yusuke<br />
Seko et al. (pp. 174–182), revealed two<br />
groups which could also be distinguished<br />
by the pigmentation of the appendages<br />
to the ascomata; E. syringiae occurred<br />
only on Syringa and probably evolved in<br />
North America, and E. ligustri and E.<br />
syringae-japonicae on Ligustrum and Syringa<br />
respectively, with the last species having<br />
arisen in eastern Asia.<br />
Three contributions are on biological<br />
and development aspects. Roger Cook et<br />
al. (pp. 183–197) report on appressorium<br />
formation on the germ tubes of 36 Erysiphe<br />
species. Unlobed appressoria (“alobatustype”)<br />
occurred in three species, while<br />
in others they were lobed. Viewed from<br />
below in the plane in contact with the<br />
host, five-lobed appressoria were formed by<br />
120° dichotomous branchings; species of<br />
Neoerysiphe and Phyllactina branched at the<br />
same angle. I found the light and scanning<br />
electron micrographs and explanatory<br />
drawings particularly illuminating. In<br />
Oidium neolycopersici, Yoshihiro Takikawa<br />
et al. (pp. 198–203) endeavoured to fund<br />
whether the point of initiation of germ<br />
tubes was triggered by contact with the<br />
host leaves or if it was predetermined; in<br />
their experiments using electrostatic spore<br />
collectors and different substrates, they<br />
showed germination was always stimulated<br />
by contact but that they almost exclusively<br />
arose subterminally regardless of the actual<br />
point of contact. Takikawa et al. (pp.<br />
204–209) also studied the behaviour of<br />
conidia in E. trifoliorum. In that species, on<br />
host and non-host leaves as well as several<br />
artificial surfaces; they discovered that the<br />
so-called “two-step” germination process<br />
was actually the result of a first unsuccessful<br />
attempt at host penetration. Microcyclic<br />
conidiogenesis, the formation of conidia<br />
directly from a conidium without little<br />
or no hyphal growth, is documented by<br />
Alexandra Pintye et al. (pp. 213–216)<br />
from species of four genera based on light<br />
and low-temperature scanning electron<br />
microscopy.<br />
Finally, Uwe Braun (pp. 210–212)<br />
provides a brief overview of the current state<br />
of systematics in these fungi, which includes<br />
a synopsis of the tribe and subtribe system<br />
to be used in the forthcoming Manual of<br />
the Erysiphales (Powdery Mildews) he has<br />
prepared with Roger Cook and which is<br />
expected to be published in April 2012;<br />
that work will evidently recognize about<br />
820 species, a marked increase from the 515<br />
accepted in Uwe’s 1987 monograph – it will<br />
be a “must have” for both field mycologists<br />
and plant pathologists worldwide.<br />
RESEARCH NEWS<br />
volume 2 · no. 2<br />
(61)
BOOK NEWS<br />
21 st Century Guidebook to Fungi. By David Moore, Geoffrey D. Robson & Anthony P. J.<br />
Trinci. 2011. ISBN 978-1-107-00676-8 (hdbk), 978-0-521-18695-7 (pbk). Pp. xii + 627,<br />
illustr., CD. Cambridge University Press, Cambridge, UK. Price £ 80, US$ 135 (hdbk),<br />
£ 40, US$ 65 (pbk).<br />
A modern mycological textbook for use in<br />
advanced university courses has been much<br />
needed, as there have already been so many<br />
advances in our understanding of their<br />
genetics, development, and relationships<br />
this century. This book grew out of courses<br />
the authors have taught at the University<br />
of Manchester for many years. It aims to<br />
capture the excitement, and present a new<br />
look, of fungi as they are now understood in<br />
a “systems biology” framework, emphasizing<br />
interactions with other organisms, adopting<br />
an integrated rather than a reductionist<br />
approach, utilizing modelling and<br />
bioinformatics when appropriate - and<br />
enhancing accessibility by an accompanying<br />
CD not just with the entire text, but also<br />
hyperlinks to both key websites and cited<br />
references.<br />
The 18 chapters are organized into six<br />
parts: Nature and origin of fungi; Fungal<br />
cell biology; Fungal genetics and diversity;<br />
Biochemistry and developmental biology of<br />
fungi; Fungi as saprotrophs, symbionts and<br />
pathogens; and Fungal biotechnology and<br />
bioinformatics. All are well-illustrated by line<br />
drawings, half-tones, tables, and “resource<br />
boxes”, and are accompanied by a list of<br />
“references and further reading”. Coloured<br />
versions of the photographs are provided<br />
in two bound-in signatures and on the CD,<br />
though the quality of the printed ones is not<br />
optimal. Also, while there is a note to say in<br />
the main text if an illustration is also in the<br />
coloured sections, no precise indication is<br />
provided which hinders their location.<br />
While most points that might be<br />
expected to be in a modern text of fungi<br />
are there, the balance of coverage reflects<br />
the research interests of the authors, and<br />
also the types of courses where fungi might<br />
still constitute a unit in a UK university<br />
today. More attention is consequently<br />
accorded to developmental, physiological,<br />
and industrial aspects (including massculture<br />
and fermenter design), for example,<br />
rather than classification, biogeography, and<br />
community ecology. However, although the<br />
overview of the fungi and their classification<br />
only have 40 pages in the main body of<br />
the book, and much of that occupied by<br />
photographs, there is an Appendix which<br />
includes descriptions down to order with<br />
example genera indicated. On the ecological<br />
side, it is great on ecosytem processes, but<br />
has little on community ecology, and no<br />
section on biogeography. The coverage of<br />
uses of fungi is particularly extensive and<br />
informative, and also well-illustrated, but<br />
surprisingly there was no section devoted to<br />
biodeterioration and challenge-testing which<br />
are so important in manufacturing. Student<br />
appeal might be enhanced by some perhaps<br />
rather unexpected features, such as that<br />
on the origin of the universe and of Earth,<br />
detailed treatments of cheese manufacture<br />
and ripening, and headings like “Ten ways<br />
to make a mushroom”. A detailed discussion<br />
of a Clitocybe nebularis colony arising under<br />
paving slabs at David Moore’s home garden<br />
is novel approach to the teaching of fungal<br />
development and structure, but did it really<br />
merit 10 pages of colour plates?<br />
A major innovation and “plus” for this<br />
work is the CD. The imbedded hyperlinks<br />
are not just to an extraordinary number of<br />
websites, but to all cited original papers with<br />
DOI numbers. The CD uses Firefox and I<br />
was most impressed by the speed at which<br />
links were made. While not all papers will be<br />
open-access and available for free download,<br />
at least their abstracts can be found; and a<br />
click on a book title or chapter can take you<br />
directly to the publisher’s sales page. Those<br />
fortunate to be located at institutions with<br />
electronic subscriptions to journals that are<br />
not open access, however, will quickly reach<br />
the full-texts. This ability to link students<br />
directly from a textbook to primary research<br />
papers is really exciting and sure to enthuse. It<br />
will also be a great short-cut for mycologists<br />
in general who have access to the CD. I can<br />
imagine the CD being carried around in<br />
many a student’s bag, not least as even the<br />
paperback version of the book weighs just<br />
over 1.9 kg.<br />
I particularly liked the line drawings<br />
incorporating colour explaining<br />
ultrastructural and developmental features.<br />
The quality of the colour photographs on the<br />
CD is superb, but the versions in the volume<br />
would have benefitted from being printed<br />
to higher specifications. For the systematist<br />
there are inevitably some minor irritations,<br />
for instance the indication of Pneumocystis<br />
carinii and P. jirovecii as synonyms (p. 30)<br />
when they are actually distinct species,<br />
a Coprinus that did not get changed to<br />
Coprinopsis (Fig 11.2) through most were<br />
changed where appropriate, and use of the<br />
kingdom name Chromista over Straminipila<br />
on the grounds of priority of publication,<br />
a criterion that does not apply above the<br />
rank of family. However, such small points<br />
are more than compensated for by a second<br />
Appendix which provides a splendid account<br />
of the terminology of mycological structures<br />
from mycelia to sporophores, and including<br />
conidiogenesis patterns, and types of asci,<br />
basidia, hyphae, sporophores, and t<strong>issue</strong>s.<br />
No two groups of authors are ever likely<br />
to concur about just what should go into<br />
an advanced student text, and what the<br />
balance between the various topics should<br />
be. There can be no doubt, however, that<br />
this will be great as a main text for mycology<br />
courses at advanced degree and masters<br />
levels, and also a valuable sourcebook for<br />
up-to-date information on diverse aspects<br />
for the subject. The authors can be proud of<br />
this achievement, which has the potential<br />
to enthuse and inspire a new generation of<br />
experimental mycologists. I cannot commend<br />
it too highly, and am recommending on the<br />
postgraduate courses to which I contribute.<br />
(62) ima fUNGUS
Evolution of Fungi and Fungal-like Organisms. Edited by Stefanie Pöggeler &<br />
Johannes Wöstemeyer. 2011. ISBN 978-3-642-19973-8. Pp. xix + 345, figs 60, col. 10.<br />
Heidelberg, Germany: Springer. [The Mycota Vol. XIV. Edited by Karl Esser.] Price<br />
£ 180, 199.95 €, US$ 269.<br />
There have been so many insights into the<br />
evolution of fungi since the two Systematics<br />
and Evolution books in The Mycota (Vol.<br />
VII A & B, McLaughlin DJ, McLaughin<br />
EG, Lemke PA, eds, 2001), that a volume<br />
addressing this topic was a natural extension<br />
of the series. Essentially, this is a mixture of<br />
12 in-depth topical and scholarly reviews<br />
on different aspects of an enormous topic<br />
arranged in four groups.<br />
The first concern the ‘Evolutionary<br />
roots of fungi’, where there is a detailed<br />
and welcome discussion of the position<br />
of the Fungi within Opisthokonta (Carr &<br />
Baldaup) which they accept as including<br />
Rozella – which is now placed in phylum<br />
Cryptomycota (see <strong>IMA</strong> <strong>Fungus</strong> 2: 173–175,<br />
2011). Opisthokonts other than Fungi<br />
are referred to as Holozoa, and as sister<br />
to Fungi and also outside Holozoa are a<br />
group of amoeboid protists which were<br />
unfamiliar to me (Nucleariida) and are yet<br />
to be included in multigene phylogenies<br />
along with microsporidians and other<br />
Fungi. The situation with Microsporidia is<br />
lucidly explained by Williams & Keeling,<br />
who consider that these mitochondriondeficient<br />
organisms may have acquired<br />
an ADP/ATP transferase by horizontal<br />
gene transfer from bacteria which enables<br />
them to exploit ATP in animal hosts. The<br />
impact of environmental DNA analyses<br />
on the concept of the fungal kingdom is<br />
discussed by Jones & Richards; they explain<br />
gene-library and 454 amplicon sequencing<br />
approaches and report in particular on<br />
their studies of sequences from marine and<br />
freshwater habitats and the discovery and<br />
visualization of Cryptomycota.<br />
Three chapters are included under<br />
‘Evolution of signalling in fungi and fungallike<br />
organisms’. The phrase ‘fungal-like’ is<br />
particularly appropriate as the first concerns<br />
dictyostelids, which are now placed in<br />
the amoebozoans as a sister group to the<br />
opisthokonts. Dictyostelium discoideum has<br />
been the subject of penetrating studies since<br />
Kenneth B. Raper started to investigate<br />
it in the 1940s and these still make the<br />
pages of Nature today. Schaap provides a<br />
review which also covers the relationships<br />
of social amoebae, and a diagrammatic<br />
phylogenetic tree showing the evolution<br />
of 20 morphological features which many<br />
working on true fungi will wish they could<br />
emulate for ‘their’ model taxa. Studies<br />
on Saccharomyces cerevisae surely most<br />
closely approach those on Dictyostelium,<br />
and Pöggeler reviews the occurrence and<br />
functions of pheromones and pheromone<br />
receptors in both this yeast and diverse<br />
filamentous ascomycetes where pheromone<br />
genes have also been found in both homoand<br />
heterothallic species. A particularly<br />
detailed and welcome review of mating<br />
types and sexuality types in basidiomycetes<br />
by Kües, James & Heitman follows, ranging<br />
from rusts and smuts to different orders<br />
of tremelloid fungi and agarics; the new<br />
term ‘unipolar’ is coined for homothallic<br />
reproduction involving a single mating type<br />
producing meiotic progeny.<br />
The largest section concerns the<br />
‘Evolution of mutualistic systems and<br />
metabolism in fungi’. Schüßler & Walker,<br />
trace the origins of Glomeromycota from the<br />
earliest fossils > 460 Myr ago and discuss<br />
their role in the evolution of land plants<br />
as well as providing an overview of the<br />
taxonomy of the group. Schmitt addresses<br />
sporophore evolution in ascomycetes,<br />
including lichenized groups, and the<br />
now well-established development of<br />
similar ascomata in diverse orders and<br />
classes – but not the currently contentious<br />
<strong>issue</strong> of whether the earliest filamentous<br />
ascomycetes were lichenized and subsequent<br />
loss of the ability to form lichens. The results<br />
of genomic analysis in Dothideomycetes<br />
are reviewed by Hane, Williams & Oliver<br />
now data on three representatives are<br />
available, all plant pathogens (Leptosphaeria<br />
maculans, Mycosphaerella graminicola,<br />
and Phaeosphaeria nodorum); the extent<br />
of the data is impressive, and includes<br />
mitochondrial DNA and evidence of<br />
horizontal transfer of pathogenicity genes.<br />
The last two contributions in this section<br />
concern the evolution of particular groups<br />
of genes or gene families. Those involved<br />
in ‘secondary metabolism’ are treated<br />
by Teichert & Nowrousian, principally<br />
polyketide synthase (pks) genes, but also<br />
covered are peptides, alkaloids, terpenes and<br />
melanins – the latter being treated in more<br />
depth than I have noted elsewhere. Genes<br />
involved in carbonic anhydrase enzyme<br />
production in fungal-like as well as fungal<br />
organisms are reviewed by Elleuche; these<br />
encode five unrelated classes of enzymes,<br />
and plant-type β-carbonic anhydrases in<br />
filamentous ascomycetes is linked to gene<br />
duplication following separation from<br />
yeasts.<br />
The final section, ‘Evolutionary<br />
mechanisms and trends’, has just two<br />
chapters. The first, by Whittle &<br />
Johannesson, returns to the evolution of<br />
mating-type loci and chromosomes, but<br />
this time in Neurospora tetrasperma – and<br />
so might have been placed in the second<br />
section of the book. In a similar way,<br />
the second, on the evolution of special<br />
metabolisms by Schimek, could have been<br />
placed in the third group with that covering<br />
pks genes. It is, however, much more wideranging<br />
in the range of compounds covered<br />
and serves as a comprehensive introduction<br />
to this fascinating aspect of fungi.<br />
I can appreciate that the editors had<br />
difficult choices to make in deciding<br />
what to include in a volume of this title.<br />
I was excited at the prospect of the title,<br />
BOOK NEWS<br />
volume 2 · no. 2<br />
(63)
BOOK NEWS<br />
and found some of the contributions<br />
illuminating, but was left feeling frustrated<br />
by what was not there. Examples include:<br />
the overall fossil record (including a<br />
discussion of the enigmatic gigantic<br />
Prototaxites now increasingly featured<br />
in fungal texts), the nature of the first<br />
filamentous fungi on land, the evolution of<br />
ectomycorrhizas and expansion of forests,<br />
convergent evolution in basidiomes, cospeciation<br />
with animals (including insects)<br />
and plants, biogeography and speciation<br />
through genetic drift, evolution in mitotic<br />
fungi, cryptic speciation, species concepts,<br />
etc. Perhaps some of these topics will be<br />
covered in future in the six additional<br />
volumes which Karl Esser indicates are to<br />
come in an addendum to the series Preface<br />
(p. xi)? Availability will remain a problem<br />
because of the cost, though an e-book<br />
version can also be purchased, However,<br />
I really must concur with the sentiments<br />
expressed by Frank Odds (2004) when<br />
reviewing Volume XII that “Perhaps<br />
The Mycota could form the embryo for a<br />
new review journal in mycology?” Surely<br />
that would be the best way to make such<br />
scholarly reviews more widely available.<br />
Odds F (2004) [Book review.] Medical mycology. The Mycota. XII. Human Fungal Pathogens. Mycological Research 108: 463–464.<br />
Metagenomics: current innovations and future trends. Edited by Diana Marco. 2011.<br />
ISBN 978-1-904455-87-5. Pp. xii + 296, col. pl. 1. Caister Academic Press, Norfolk, UK.<br />
Price £ 159, US$ 319.<br />
Metagenomics, the comparison of entire<br />
communities from the genomes of the<br />
constituent organisms, is exciting cuttingedge<br />
research with respect to the exploration<br />
of phylogenetic and functional genetic<br />
diversity in particular ecological niches. The<br />
approach is very much technique-driven,<br />
as possibilities expand as instrumentation<br />
and protocols develop. So far, the majority<br />
of studies have focussed on bacterial<br />
groups and viruses, but the approach is<br />
increasingly being taken up by fungal<br />
ecologists fortunate to have access to the<br />
necessary equipment – and generating some<br />
illuminating results, especially with respect<br />
to phylogenetic diversity.<br />
In entering a new field or using a new<br />
technology, topical syntheses can be viewed<br />
as equivalent to a crash-course. This text has<br />
15 chapters, only one developed to fungi<br />
(see below), with the rest centred on either<br />
techniques or bacterial groups or viruses<br />
in particular communities. The value of<br />
this text for mycologists is that in presents<br />
state-of.-the art information on the methods<br />
and their limitations, and has examples of<br />
actual applications in other groups that<br />
might be mirrored in future mycological<br />
investigations.<br />
The chapter on current approaches<br />
(Meiring et al., pp. 1–19) is especially<br />
informative, explaining the theory<br />
and practice of both sequence-driven<br />
and function-driven approaches and<br />
how they relate to the complementary<br />
metaproteomics and metabolomics. There<br />
is a separate chapter on next-generation<br />
sequencing and the use of 454 and other<br />
sophisticated machines (Walshaw et al.,<br />
pp. 63–88) which I found to be a most<br />
illuminating introduction to the protocols<br />
and procedure of this much talked-about<br />
methodology. That on microarrays (van<br />
Norstrand et al., 265–288) was similarly<br />
most helpful, and includes a section on<br />
GeoChips (functional gene arrays) which<br />
were new to me. Other chapters cover<br />
topics such as bacterial genealogy, viruses,<br />
the human microbiome, sequencing from<br />
uncultured single cells (using flow cytometry<br />
for their separation), bioremediation, host<br />
engineering and enzyme discovery, nonfungal<br />
plant pathogens.<br />
The single fungal chapter concerns<br />
arbuscular mycorrhizal fungi, and has<br />
been prepared by Paola Bonfante´s group<br />
in Turin (Bianciotto et al., pp. 161–178).<br />
This includes an overview of all studies<br />
using metagenomic approaches to date,<br />
from the pioneering work by Alastair<br />
Fitter´s group in 1998–99). The practical<br />
relevance is clear in view of the large<br />
numbers of species that would otherwise<br />
have remained unrecognized by reliance<br />
on non-molecular approaches. By the end<br />
of September 2009, there was a staggering<br />
12 274 “uncultured” AM fungal genomes<br />
recognized for which there was no link<br />
to described morphospecies. The <strong>issue</strong> of<br />
overestimations due to artefacts arising from<br />
the fragmentation of genomes is flagged up<br />
here, and perhaps might have benefited from<br />
more discussion in some others as this is so<br />
critically important to estimates of actual<br />
organismal diversity derived.<br />
In summary, if you are a mycologist<br />
contemplating adopting, or even already<br />
using, metagenomic and next-generation<br />
sequencing technologies, this work<br />
should be consulted when designing work<br />
programmes or interpreting the mass of<br />
generated data.<br />
Los Hongos de Panamá: introducción a la identification de los macroscópicos. By<br />
Gastón Guzmán & Meike Piepenbring. 2011. ISBN 978-607-7579-21-2. Pp. xiv + 372,<br />
figs 710 (most col.), tables 3. Xalapa, Mexico: Instituto de Ecología A.C. Price: Not indicated.<br />
(64) ima fUNGUS
December 2010 was the centenary of the<br />
arrival of the first scientific expedition of<br />
the Smithsonian Institution to Panamá,<br />
and what better way to mark the event than<br />
by a splendidly illustrated work on fungi.<br />
Meike Piepenbring, who is based at the J.<br />
W. Goethe Universität in Frankfurt but<br />
also holds a position in the Universidad<br />
Autónoma de Chiriquí in Panamá, is<br />
well-known for her detailed and on-going<br />
investigations into the diversity of fungi of<br />
all kinds that occur in Panamá. Here she<br />
has joined forces with Gaston Guzmán,<br />
who has, and continues to, so energetically<br />
promote mycology in the region.<br />
It was pleasing to see a broad<br />
interpretation of macroscopic fungi<br />
adopted, encompassing not only larger<br />
basidiomycetes and discomycetes, but also<br />
a range of slime-moulds, pyrenomycetes,<br />
and lichens. In all, 226 species are treated in<br />
detail, with discussions of their microscopic<br />
as well as macroscopic features, differences<br />
from similar taxa, and references to<br />
pertinent literature; they are arranged<br />
alphabetically which facilitates their<br />
location. The colour photographs include<br />
some close-ups showing diagnostic features<br />
such as gill types, or slices through them<br />
to show the arrangement of perithecia in a<br />
stroma (that of Hypoxylon haematostroma<br />
on p. 136 is a striking example). According<br />
to the Abstract (p. ii), over 700 species are<br />
covered to some extent. There is a series<br />
of 13 keys to genera, and in some cases<br />
species, based on the gross morphology<br />
of the sporophores, an introduction<br />
including microscopic characters which<br />
is well-illustrated by clear line drawings, a<br />
glossary, taxonomic arrangement of taxa<br />
discussed, an appendix which includes<br />
further observations on 29 genera or groups<br />
of species (with especially informative<br />
discussions of several gasteroid groups),<br />
a taxonomic index which lists synonyms<br />
under treated species, and a main index<br />
including entries by species epithet.<br />
This lavishly illustrated book should<br />
do much to encourage both students and<br />
citizen scientists to take a deeper interest<br />
in fungi. While it is in Spanish, it is<br />
nevertheless easy to use by Anglophones<br />
with knowledge of a few key words and<br />
mycological terms. I just wish it had been<br />
available when I visited Barro Colorado<br />
Island in 1995, and was so fascinated by the<br />
array of Xylaria species sprouting from logs<br />
– 11 receive full treatment here, several of<br />
which I had not seen colour photographs of<br />
before. While the colour reproduction and<br />
quality of some of the photographs might<br />
have been better, and the taxonomy is in<br />
some cases conservative (e.g. the retention of<br />
Coprinus for C. cinereus and C. stercoreus),<br />
this production represents a tremendous<br />
achievement of which the authors can be<br />
justly proud.<br />
The production of this book was<br />
made possible through the support of the<br />
Smithsonian Tropical Research Institute<br />
(STRI) in Panamá and eight other bodies.<br />
Only 500 copies have been printed, so if you<br />
work on, or are interested in, Central and<br />
South American fungi, it would be advisable<br />
to endeavour to secure a copy soon as it is<br />
sure to prove very popular in the region.<br />
Champignons Comestibles des Forêts denses d’Afrique Centrale: taxonomie et identification.<br />
By Hugues E. Ndong, Jérôme Degreef & André De Kesel. 2011. ISSN 1784-<br />
1283 (hd copy), 1784-1291 (online PDF). Pp. viii+ 253, figs 151 (many col.), tables 1,<br />
backpocket. Brussels, Belgium: Royal Belgian Institute of Natural Sciences. [ABC Taxa<br />
Vol. 10.] Price: Free (developing countries), 15.45 € (other countries).<br />
BOOK NEWS<br />
This work is one of series aimed at<br />
accelerating taxonomic capacity building<br />
in developing countries, and the first on<br />
fungi. The aim is to provide sufficiently<br />
detailed state of the art treatments to enable<br />
recipients to embark on the taxonomy of<br />
the group treated. Consequently, almost one<br />
third of the book is devoted to background<br />
information: current knowledge,<br />
ethnomycology, preparation for a collecting<br />
trip, field and laboratory requisites,<br />
collection, documentation, photography,<br />
description, microscopic preparations,<br />
measurements, preservation, and herbarium<br />
formation. In order to facilitate detailed<br />
descriptions there is a detailed account of<br />
anatomical and morphological features<br />
illustrated by fine line drawings. A great<br />
deal of care has been taken to make sure<br />
a beginner would find all he or she needs<br />
to start to make contributions to the field,<br />
though a section on nomenclature could<br />
have been a useful addition. Inside the<br />
back cover, is a pocket including a sample<br />
recording card for descriptive information,<br />
where pertinent characters can be quickly<br />
circled to facilitate the rapid processing of<br />
specimens.<br />
Accounts of 62 edible species follow,<br />
most with a text page facing a figure which<br />
includes well-executed coloured illustrations<br />
of intact sporophores and vertical sections,<br />
and also drawings of key microscopic features.<br />
In some cases photographs of fresh specimens<br />
volume 2 · no. 2<br />
(65)
BOOK NEWS<br />
are also provided. The species are drawn<br />
from a wide range of genera, amongst which<br />
are Amanita, Auricularia, Cantharellus,<br />
Cookeina, Lactarius, Marasmius, Polyporus,<br />
Russula, Schizophyllum, and Termitomyes.<br />
The text has detailed macro- and microscopic<br />
descriptive data, information on ecology<br />
and distribution, and notes discussing<br />
the separation from similar taxa – but<br />
surprisingly not on preparation or taste. A<br />
comprehensive glossary is provided, and the<br />
reference list is extensive.<br />
The authors, all who have experience<br />
of working in the region, have clearly put<br />
an immense amount of thought into the<br />
volume and it is deserves to be widely<br />
distributed in central Africa. On the back<br />
cover, Bart Buyck states that the book<br />
should occupy prime position in the<br />
libraries of mycologists, particularly for the<br />
full introduction and practical approach;<br />
I totally concur with his sentiments.<br />
The concept of making this work freely<br />
available in developing countries can only<br />
be applauded and was made possible by<br />
the foresight of the Belgian Development<br />
Cooperation. That is an aspect of capacity<br />
building which is too often overlooked in<br />
aid programmes. My one sadness is that<br />
the text is entirely in French, as an English<br />
translation would be so valuable for use<br />
in the non-Francophone countries in the<br />
region; it would be tremendous if additional<br />
funds to do that could be found.<br />
Love, Sex and Mushrooms: adventures of a woman in science. By Cardy Raper. 2011.<br />
ISBN 978-0-615-43440-7. Pp. xii + 254, illustr. Burlington, VE: C. Raper. Price: US$<br />
18.95.<br />
This is a very personal, and in parts emotive,<br />
account of aspects of both the personal and<br />
professional life of Cardy Raper, the wife<br />
of the renowned fungal geneticist John<br />
R. Raper (1911 –1974) 1 and well-known<br />
for his monograph on the genetics of<br />
sexuality in macromycetes (Raper 1966).<br />
She recounts the trials she has experienced<br />
in establishing herself as a respected<br />
fungal geneticist in her own right, in an<br />
academic climate which often seemed to<br />
conspire against female scientists. She also<br />
documents the tedious and often frustrating<br />
attempts to obtains samples of hormones<br />
involved in the sexual behaviour of Achlya,<br />
and to obtain isolates of elusive mating<br />
types before their work changed its focus<br />
to Schizophyllum commune – a change in<br />
course largely due to graduate student Haig<br />
Papazian arriving from London. There are<br />
insights into her experiences as a faculty<br />
wife in the USA and as a mother, and into<br />
her personal life and relationship problems<br />
that she had to contend with. There are also,<br />
perhaps sometimes tongue-in-cheek, asides<br />
about other mycological luminaries she<br />
encountered or who influenced her, such<br />
as Karl Esser and Dirk Wessels. The actual<br />
science is not described in detail, however,<br />
though there are tantalizing tid-bits here<br />
and there; not being a fungal geneticist, I<br />
would have found citations and discussions<br />
of actual papers illuminating (those by<br />
Cardy are, however, listed on her website,<br />
http://cardyraper.com).<br />
Cardy comes over as a person determined<br />
to succeed and full of enthusiasm for her<br />
chosen area of science. This autobiography<br />
demonstrates that a scientific path can be<br />
long and hard, and that is something as<br />
true now as it was during Cardy’s struggling<br />
years – especially when tenure is now so<br />
difficult to secure in many countries. The<br />
preliminary pages include comments from<br />
two distinguished fungal geneticists, Lorna<br />
Casselton and Peter Day. Lorna’s ends with<br />
the phrase “This is the personal account<br />
of an exceptional scientist”, and Peter’s<br />
with the remark that she lived up to the<br />
slogan of a Harvard band, “Illegitimum<br />
non carborundrum: don’t let the bastards<br />
grind you down”. It could be seen either as a<br />
cautionary tale or as an inspiration to aspiring<br />
researchers, and of course will also be of<br />
interest to those wishing to know more of<br />
both John and Cardy’s backgrounds.<br />
1<br />
Brother of Kenneth B. Raper (1908–1987).<br />
Raper JR (1966) Genetics of Sexuality in Higher Fungi. New York: Ronald Press.<br />
(66) ima fUNGUS
FORTHCOMING MEETINGS<br />
International and regional meetings which are entirely mycological or have a mycological content<br />
2012<br />
International Association for Lichenology (IAL7) – Lichens: from genome to ecosystems in a changing world<br />
9–13 January 2012<br />
Bangkok, Thailand<br />
Contact: Lichen Research Unit, Ramkhamhaeng University; e-mail: lichen.ial7@gmail.com<br />
<br />
Fungal Pathogens: from basic biology to drug discovery [A Keystone Symposium]<br />
15–20 January 2012<br />
Community Convention Center, Santa Fe, New Mexico, USA<br />
<br />
FORTHCOMING MEETINGS<br />
5 th Advances Against Aspergillosis<br />
26–28 January 2012<br />
Lutfi Kirdar Convention and Exhibition Centre, Istanbul, Turkey<br />
Contact: Hartley Taylor Medical Communications, Henderson House, New Road, Princess Risborough, Buckinghamshire HP27 0JN, UK;<br />
derry@hartleytaylor.co.uk<br />
<br />
International Conference on Mycology and Plant Pathology: Biotechnological Approaches<br />
27–29 February 2012<br />
Baranas Hindu University, Varanai, India<br />
Contact: Dr Ravindra N. Kharwar, Centre for Advanced Studies in Botany, Baranas Hindu University, Varanai, 221005 India; rnkharwar@<br />
gmail.com or icmpb2012@gmail.com<br />
<br />
11 th European Congress of Fungal Genetics<br />
30 March–2 April 2012<br />
Phillips-Universität Marburg, Germany<br />
Contact: Congress Office, c/o Intelligent Events, 126 High Street, Dunblane, Perthsire FK15 0ER, UK; contact@ecfg.info<br />
<br />
CBS Symposium: One <strong>Fungus</strong> = Which Name?<br />
[In collaboration with the Nomenclature Committee for Fungi and the International Commission on the Taxonomy of Fungi]<br />
12–13 April 2012<br />
Amsterdam, The Netherlands<br />
Contact: Pedro Crous, CBS-KNAW Fungal Biodiversity Centre, PO Box 85167, 3508 AD Utrecht, The Netherlands; p.crous@cbs.knaw.nl<br />
<br />
18 th International Society for Human and Animal Mycology (ISHAM)<br />
11–15 June 2012<br />
Berlin, Germany<br />
<br />
2 nd Annual International Symposium on Mycology (ISM-2012)<br />
30 July–1 August 2012<br />
Guangzhou, China<br />
Contact: Maya Chen, East Area F11 Building 1, Dalian Ascendas IT Park, 1 Hui Xian Yuan, Dalian Hi-Tech Industrial Zone, LN 116025,<br />
China; maya@bitconferences.com<br />
<br />
volume 2 · no. 2<br />
(67)
FORTHCOMING MEETINGS<br />
Fungal Interactions [organized with the British Mycological Society]<br />
3–6 September 2012<br />
Alicante University, Spain<br />
<br />
2 nd Annual World Congress on Marine Biotechnology (WCMB-2012)<br />
20–23 September 2012<br />
World Expo Center, Dalian, China<br />
Contact: Doris Han, East Area F11 Building 1, Dalian Ascendas IT Park, 1 Hui Xian Yuan, Dalian Hi-Tech Industrial Zone, LN 116025,<br />
China; doris@bit-ibio.com<br />
<br />
2013<br />
10 th International Congress of Plant Pathology<br />
25–31 August 2013<br />
Beijing, China<br />
<br />
2014<br />
10 th International Mycological Congress (IMC10)<br />
3–8 August 2014<br />
Bangkok Convention Center, Bangkok, Thailand<br />
Contact: Lekha Manoch; e-mail: agrlkm@ku.ac.th<br />
IUMS XIV International Congress of Mycology<br />
[with the Congresses of Bacteriology and Applied Microbiology, and also Virology]<br />
27 July–1 August 2014<br />
Montreal, Canada<br />
Contact: e-mail: iums3014@nrc-cnrc.gc.ca<br />
<br />
NOTICE<br />
<strong>IMA</strong> <strong>Fungus</strong> is compiled and edited by David L. Hawksworth (Facultad de Farmacia, Universidad Complutense de Madrid) on<br />
behalf of the Executive Committee of the International Mycological Association. All unsigned items in the journal may be<br />
attributed to, him.<br />
Items for consideration for inclusion in all sections of the journal should be submitted to David at d.hawksworth@nhm.ac.uk.<br />
Books for possible coverage in the Book News section should be mailed to: <strong>IMA</strong> <strong>Fungus</strong>, Milford House, The Mead, Ashtead,<br />
Surrey KT21 2LZ, UK.<br />
(68) ima fUNGUS
doi:10.5598/imafungus.2011.02.02.01 <strong>IMA</strong> <strong>Fungus</strong> · volume 2 · no 2: 113–120<br />
One <strong>Fungus</strong> = One Name: DNA and fungal nomenclature twenty years after<br />
PCR<br />
John W. Taylor<br />
ARTICLE<br />
University of California Berkeley, 111 Koshland Hall, Berkeley, CA 94720-3102, USA; e-mail: jtaylor@berkeley.edu<br />
Abstract: Some fungi with pleomorphic life-cycles still bear two names despite more than 20 years of molecular<br />
phylogenetics that have shown how to merge the two systems of classification, the asexual “Deuteromycota”<br />
and the sexual “Eumycota”. Mycologists have begun to flout nomenclatorial regulations and use just one name<br />
for one fungus. The International Code of Botanical Nomenclature (ICBN) must change to accommodate<br />
current practice or become irrelevant. The fundamental difference in the size of fungi and plants had a role in<br />
the origin of dual nomenclature and continues to hinder the development of an ICBN that fully accommodates<br />
microscopic fungi. A nomenclatorial crisis also looms due to environmental sequencing, which suggests that<br />
most fungi will have to be named without a physical specimen. Mycology may need to break from the ICBN<br />
and create a MycoCode to account for fungi known only from environmental nucleic acid sequence (i.e. ENAS<br />
fungi).<br />
Key words:<br />
Amsterdam Declaration<br />
ENAS<br />
MycoCode<br />
nomenclature<br />
pleomorphic fungi<br />
Article info: Submitted: 8 June 2011; Accepted: 15 June 2011; Published: 12 July 2011.<br />
INTRODUCTION<br />
It has been a bit over two decades since the polymerase chain<br />
reaction (PCR) changed evolutionary biology in general and<br />
fungal systematics in particular. Even before PCR became<br />
generally available, mycologists realized that the evolutionary<br />
record contained in the nucleic acid sequence of every fungus<br />
could be used to merge two systems of nomenclature that<br />
had been employed in most fungi, i. e. one for the “Eumycota”<br />
based on sexual morphology and the “Deuteromycota” based<br />
on all other morphologies (Berbee & Taylor 1992, Bruns et al.<br />
1991, Guadet et al. 1989, Reynolds & Taylor 1992). Why, then,<br />
has it taken more than two decades for nomenclature to catch<br />
up with biology, and why is the possibility of nomenclatorial<br />
rapprochement now being taken seriously? These questions,<br />
and three others posed to the participants in this symposium<br />
will be the subject of this contribution: Does DNA sequencing<br />
make dual nomenclature superfluous? Can the International<br />
Code of Botanical Nomenclature (ICBN) (McNeill et al. 2006)<br />
be modified to enable this process, or would a MycoCode<br />
be more effective? How can the mycological community get<br />
rid of the legacy of dual nomenclature and Article 59 without<br />
nomenclatural chaos?<br />
Two examples illustrate the practical problems raised<br />
by dual nomenclature. First, this year, while serving as a<br />
member of a governmental committee researching the<br />
use of mycoherbicides to eradicate drug crops, it fell to me<br />
to explain the nomenclature of two poppy pathogens that<br />
are sister species, one named as a teleomorph Crivellia<br />
papaveracea and the other as an anamorph, Brachycladium<br />
papaveris (Inderbitzin et al. 2006) (Fig. 1). The fifteen other<br />
members of the committee, eleven academics and four very<br />
knowledgeable staff, stared at me in disbelief when I said that<br />
sister species could have different generic names. Second,<br />
together with Tom Bruns, I have been directing research<br />
about fungi that naturally decay plants proposed as sources<br />
of lignocellulose for the production of biofuels. In the course<br />
of this work, we have sequenced ITS using DNA isolated<br />
from the decaying grasses and compared the sequences to<br />
those deposited in GenBank. Often, a single sequence will be<br />
attached to two names; you guessed it, it’s the same fungus<br />
with some GenBank sequences having been deposited<br />
under the teleomorph name and others under the anamorph<br />
name. Perpetuation of dual nomenclature when we have the<br />
means to abandon it is hindering mycology, both scientifically<br />
and socially.<br />
Dual nomenclature has persisted for the past 20 years<br />
because few mycologists are deeply interested in both<br />
molecular phylogenetics and nomenclature. One <strong>Fungus</strong><br />
= One Name has gained momentum, as evidenced by this<br />
conference, because mycologists who are studying the<br />
Dedication: Dedicated to Don Reynolds, mycological iconoclast,<br />
whose sabbatical visit to Berkeley from the Los Angeles<br />
County Museum of Natural History more than 20 years ago<br />
stimulated thought about One <strong>Fungus</strong> = One Name.<br />
© 2011 International Mycological Association<br />
You are free to share - to copy, distribute and transmit the work, under the following conditions:<br />
Attribution:<br />
You must attribute the work in the manner specified by the author or licensor (but not in any way that suggests that they endorse you or your use of the work).<br />
Non-commercial: You may not use this work for commercial purposes.<br />
No derivative works: You may not alter, transform, or build upon this work.<br />
For any reuse or distribution, you must make clear to others the license terms of this work, which can be found at http://creativecommons.org/licenses/by-nc-nd/3.0/legalcode. Any of the above conditions can be waived if you get<br />
permission from the copyright holder. Nothing in this license impairs or restricts the author’s moral rights.<br />
volume 2 · no. 2 113
John W. Taylor<br />
ARTICLE<br />
Pleospora<br />
Alternaria<br />
Crivellia papaveracea/Brachycladium penicillatum<br />
Crivellia sp (unnamed)/Brachycladium papaveris<br />
Fig. 1. Phylogenetic relationships of the sister species Crivellia papaveracea<br />
and Brachycladium papaveris, the former named as teleomorphic<br />
and the latter as an anamorphic fungus. The Crivellia state<br />
of B. papaveris remains unnamed due to a lack of suitable material to<br />
serve as a nomenclatural type (Inderbitzen et al. 2006).<br />
molecular phylogenetics of economically important fungal<br />
groups have begun naming newly recognized genus-level<br />
clades with just one Ascomycota name, whether or not<br />
the fungus exhibits sexual reproduction. The first thorough<br />
exploration of this practice was provided by Crous et al.<br />
(2006), whose revision of the Botrysphaeriaceae includes<br />
this sentence, “Separate teleomorph and anamorph names<br />
are not provided for newly introduced genera, even where<br />
both morphs are known.” Where a teleomorph name was<br />
available, as in the case of Botryosphaeria, the authors used<br />
it. Where only anamorph names were available, they were<br />
used, e.g. Macrophomina or Neoscytalidium. Where a new<br />
clade was segregated from an existing teleomorph genus,<br />
and best distinguished by the anamorphic morphology, the<br />
new name reflected the anamorph, e.g. Neofusicoccum.<br />
Matters were taken further in a study of Penicillium species<br />
by Houbraken et al. (2010). As they put it, “Using this approach<br />
and applying the concept of one name for one fungus (Reynolds<br />
& Taylor 1992), we have chosen to describe these two<br />
species under [their] anamorphic name.” That is, Houbraken<br />
et al. described new species that have both anamorphic and<br />
teleomorphic states as species in the anamorph-typified genus<br />
Penicillium and ignored the existing teleomorphic generic<br />
name, Eupenicillium. These actions are clearly outside the<br />
ICBN and constitute a social rebellion. Though smaller and far<br />
less important than social rebellions concerning, for example,<br />
women’s rights, the rights of African Americans, or those of<br />
homosexuals, this mycological rebellion is similar to the others<br />
in that activism has outpaced the law and the law must now<br />
change or become irrelevant.<br />
Dual nomenclature has a long history. The choice made<br />
by Houbraken et al. ( 2010) to use the anamorph name<br />
Penicillium over the teleomorph name Eupenicillium for one<br />
of the most economically important fungi echoes the choice<br />
made more than 40 years earlier by Raper & Fennel (1965)<br />
when they applied the anamorphic name Aspergillus to all<br />
members of that genus whether or not the species also<br />
produced a sexual structure. Forty years are not enough to<br />
understand the origins of dual nomenclature, to do that we<br />
have to go all the way back to Linnaeus and the beginning<br />
of botanical nomenclature. In this tour back through time, our<br />
guides will be Weresub &Pirozynski through their excellent<br />
article on the history of fungi that produce both meiotic<br />
and mitotic spores, that is, pleomorphic fungi (Weresub<br />
& Pirozynski 1979) and the opening chapters of Selecta<br />
Fungorum Carpologia, the monumental work of Louis-René<br />
Tulasne and Charles Tulasne (Fig. 2) (Tulasne & Tulasne<br />
1861).<br />
The Tulasne’s point out that Linnaeus based his plant<br />
taxonomy on floral morphology and that he could demonstrate<br />
that each plant had but one type of flower. At a time when fungi<br />
were considered to be plants, and fungal spores were equated<br />
with seeds, Linnaeus extended his taxonomic concept to fungi.<br />
The Tulasne brothers then argue that Linnaeus had such an<br />
influence over his mycological contemporaries, Fries foremost<br />
among them, that these mycologists were in denial about<br />
pleomorphy, despite their being able to see more than one<br />
type of “seed” through their lenses.<br />
“In the Mucedinei [Fries] sees the conidia . . . but everywhere<br />
he flatly denies that there occur “two kinds of sporidia on the<br />
same plant”, exactly as if he had heard, sounding in his ears,<br />
the loud voice of Linnaeus, crying “ It would be a remarkable<br />
doctrine – that there could exist races differing in fructification,<br />
but possessing one and the same nature and power; that one<br />
and he same race could have different fructifications; for the<br />
basis of fructification, which is also the basis of all botanical<br />
science, would thereby be destroyed, and the natural classes<br />
of plants would be broken up” (Tulasne & Tulasne 1861: 48 1 ).<br />
The brothers go on to chide Linnaeus, adding “But since<br />
the illustrious author always completely abjured the use<br />
of magnifying glasses, and therefore scarcely ever tried to<br />
describe accurately either conidia or spores, we fear (may he<br />
pardon the statement) that he really knew very few seeds of<br />
either kind” (Tulasne & Tulasne 1861: 48-49). The influence<br />
that the size of an organism has on its systematics can be<br />
profound (Taylor et al. 2006). The fact that the overwhelming<br />
majority of plants are macroscopic while the overwhelming<br />
majority of fungi are microscopic still affects nomenclature and<br />
will be revisited near the end of this article.<br />
Louis René and Charles Tulasne went on to argue<br />
against mycological denial of pleomorphy when they wrote,<br />
“The fungus upon which we are now touching [Pleospora] is<br />
not only almost the commonest of all belonging to its order,<br />
but also affords a wonderful proof of our doctrine concerning<br />
the multiple nature of the seeds of species of fungi“ (Tulasne<br />
& Tulasne 1861: 248). One cannot help wondering if the<br />
brothers guessed not only that their work was controversial,<br />
but that the mycological world was heading toward dual<br />
nomenclature, when they wrote, “As today we have seen<br />
the various members of the same species now unwisely torn<br />
from one another against the laws of nature . . .” (Tulasne &<br />
Tulasne 1861: 189).<br />
Alas, when the most useful characters that could be used<br />
for classification were meiosporic, and when many fungi<br />
did not exhibit them, there were not many options and the<br />
one that prevailed was dual nomenclature. Fuckel, a retired<br />
1<br />
The English translations are from the 1931 Clarendon Press<br />
(Oxford) edition, and were prepared by W B Grove and edited by A H<br />
R Buller and C L Shear.<br />
114 <br />
ima fUNGUS
One <strong>Fungus</strong> = One Name: DNA and fungal nomenclature twenty years after PCR<br />
ARTICLE<br />
Fig. 2. Louis Renè Tulasne (l) and Charles Tulasne (r). Photo: courtesy of the National Museum of Natural History, Paris.<br />
pharmacist, got the ball rolling (Fuckel 1870) and Saccardo<br />
did the heavy lifting with his Sylloge Fungorum beginning in<br />
1882 (Saccardo 1882). By 1910, the International Rules of<br />
Botanical Nomenclature (Briquet 1912) contained a section<br />
of Article 49, Art 49 bis (the precursor of the current Article 59),<br />
that forbade “botanical” names for any but the sexual stage<br />
of pleomorphic fungi and that is where matters rest with the<br />
current ICBN.<br />
Saccardo’s use of mature anamorph morphology is<br />
wonderfully convenient for classification and identification<br />
but, obviously, it is not based on evolutionary relationships.<br />
The hope that study of mitospore development would lead to<br />
a separate systematics based on evolutionary relationships<br />
began with Vuillemin (1910a, b) and Mason (1933, 1937) and<br />
led to the work of Hughes (1953), Tubaki (1958) and Barron<br />
(1968). Elegant microscopic studies of mitospore development<br />
followed (Cole & Samson 1979) and the movement reached<br />
its zenith at the second Kananaskis conference (Kendrick<br />
1979). Just as these studies of development were peaking,<br />
two events occurred in the realms of evolution and systematics<br />
that promised the irresistible appeal of a new approach and<br />
a seemingly endless supply of characters – cladistic analysis<br />
(Hennig 1966) and access to nucleic acid variation.<br />
The first applications of nucleic acid variation to fungal<br />
systematics involved DNA-DNA hybridization of yeasts<br />
(Kurtzman 1980) and then sequencing of nucleic acids.<br />
Pioneering work with painfully difficult RNA sequencing<br />
modeled on the work of bacteriologists (Walker & Doolittle<br />
1982, 1983) was followed by DNA sequencing (Gottschalk<br />
& Blanz 1984, Guadet et al. 1989, Gueho et al. 1989). But<br />
it was the discovery of the polymerase chain reaction (PCR)<br />
(Rabinow 1996, Saiki et al. 1988) that made possible the<br />
broad studies we now take for granted.<br />
The first application of PCR amplified DNA sequence<br />
to fungal phylogenetics demonstrated the evolution of<br />
hypogeous fungi from mushroom ancestors (Bruns et<br />
al. 1989; Fig. 3). This work relied on the development of<br />
primers designed to amplify regions of both mitochondrial<br />
and nuclear rDNA including the nuclear small subunit, large<br />
subunit and internal transcribed spacer (ITS), which were<br />
published the following year and have been cited a bit more<br />
often than once-a-day since then (White et al. 1990; Fig. 4).<br />
Boletus<br />
Boletus<br />
Suillus<br />
Suillus<br />
Rhizopogon<br />
Rhizopogon<br />
Fig. 3. Phylogenetic analysis of PCR amplified rDNA showing the<br />
evolution of hypogeous Basidiomycota in the genus Rhizopogon,<br />
from mushroom ancestors in the genus Suillus (Bruns et al. 1989).<br />
Adapted from Bruns et al. (1989).<br />
volume 2 · no. 2 115
John W. Taylor<br />
ARTICLE<br />
Fig. 4. Authors of the publication of PCR primers for the amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics.<br />
Left to right: Tom Bruns, Tom White, Steve Lee, and John Taylor. Photo: taken in 2010, 20 years after the publication of White et al. (1990).<br />
Sporothrix schenckii<br />
Ophiostoma stenoceras<br />
Ophiostoma ulmi<br />
Leucostoma persoonii<br />
Neurospora crassa<br />
Fig. 5. Phylogenetic analysis of PCR amplified rDNA showing the<br />
anamorphic Sporothrix schenckii nestled within the teleomorphic genus<br />
Ophiostoma (Berbee & Taylor 1992).<br />
Within a few years, analysis of PCR amplified rDNA showed<br />
that the anamorphic Sporothrix schenckii nested within the<br />
teleomorph genus Ophiostoma (Berbee & Taylor 1992; Fig.<br />
5). This work demonstrated the integration of anamorphic<br />
and teleomorphic fungi based on DNA variation, as had<br />
earlier work on Fusarium (Guadet et al. 1989). These studies<br />
showed a separate classification for “Deuteromycota” to be<br />
superfluous.<br />
That same year, Reynolds & Taylor (1992) addressed the<br />
nomenclatural implications of using DNA variation to assess<br />
the phylogenetic relationships of fungi, writing, “The use of<br />
nucleic acid sequence allows systematists to demonstrate<br />
the phylogenetic relatedness of fungi possessing and<br />
lacking meiotically produced spores. . . . This demonstration<br />
presents a serious challenge to the separate classification<br />
of these two types of fungi and undermines the elevated<br />
position that characters associated with sexual reproduction<br />
have held in the classification of higher fungi. . . . We believe<br />
that all fungi should be classified in one system and that<br />
characters associated with sexual reproduction should be<br />
given the same weight as other characters. . . . By the broad<br />
interpretation [of Article 59] in current use, the potential for<br />
pleomorphy is assumed of all fungi and the Article is applied<br />
to all fungi. . . . With an alternative and strict interpretation<br />
however, Article 59 would apply only to fungal species that<br />
have been actually demonstrated to be pleomorphic. Under<br />
the latter interpretation, sexual, asexual, and pleomorphic<br />
fungi would be classified together and form taxa would not<br />
be necessary.”<br />
Following the Fungal Holomorph Symposium in Newport<br />
(OR, USA) to discuss nucleic acid variation and the integration<br />
of anamorphic and teleomorphic classifications (Reynolds &<br />
Taylor 1993), there have been presentations and discussions<br />
on the topic at every International Mycological Congress from<br />
116 <br />
ima fUNGUS
One <strong>Fungus</strong> = One Name: DNA and fungal nomenclature twenty years after PCR<br />
ARTICLE<br />
Environmental Nucleic Acid Sequence (ENAS)<br />
Specimen Based<br />
Both ENAS and Specimen Based<br />
Fig. 6. Graph of the Operational Taxonomic Units (OTUs) added to GenBank from 1991 to 2009 showing the increasing proportion of OTUs<br />
based only on environmental nucleic acid sequence (ENAS). Adapted from Hibbett et al. (2011).<br />
Vancouver (1994; Taylor 1995) to Edinburgh (2010; Norvell et<br />
al. 2010), leading up to the present One <strong>Fungus</strong> = One Name<br />
conference and the Amsterdam Declaration (Hawksworth et<br />
al. 2011).<br />
Nucleic acid variability has proved to be useful in other<br />
areas of fungal systematics and classification related to<br />
mitotic fungi. Beginning with the mitosporic human pathogen<br />
Coccidioides immitis, DNA variation has been used to show<br />
that anamorphic fungi recombine in nature (Burt et al. 1996),<br />
that they speciate (Koufopanou et al. 1997), and that, based<br />
only on DNA variation, they can be described in the system<br />
for Ascomycota (Fisher et al. 2002). As Fisher et al. wrote<br />
when they described a new Coccidioides species as an<br />
ascomycete, “Coccidioides posadasii is morphologically<br />
indistinguishable from Coccidioides immitis. C. posadasii<br />
is diagnosed by the following nucleotide characters (given<br />
as the gene, the nucleotide position in the gene, and,<br />
parenthetically, the nucleotide fixed in C. posadasii) showing<br />
reciprocal fixation between C. immitis and C. posadasii:<br />
Chitin synthase positions 192 (A), 288 (T); Dioxygenase<br />
positions 872 (C), 1005 (C), 1020 (G), 1179 (C), 1272 (T);<br />
etc.” Of course, description is not the same as acceptance.<br />
In the case of Coccidioides posadasii, acceptance for this<br />
“Select Agent” came from an unexpected quarter, the United<br />
States Congress (Federal Register 2005).<br />
Another point made soon after PCR became available<br />
was that DNA, or even a DNA sequence, could act as the<br />
type element in a species description (Reynolds & Taylor<br />
1991). This observation has gained importance due to the<br />
advent of environmental sequencing, where mycologists use<br />
PCR primers for rDNA to amplify variable regions from DNA<br />
isolated from soil or plants. Environmental sequencing has<br />
begun to produce large numbers of rDNA sequences that<br />
document the existence of fungi for which there is neither a<br />
specimen nor a culture. Most importantly, ecological studes<br />
have shown that the number of these DNA-only fungi,<br />
or “Environmental Nucleic Acid Sequences” (ENAS) can<br />
exceed the number of fungi for which there is a culture or<br />
specimen (Jumpponen & Jones 2009, 2010). This imbalance<br />
poses a challenge to fungal classification and nomenclature<br />
that may dwarf the challenge of integrating anamorphic and<br />
teleomorphic fungi.<br />
David Hibbett, in his plenary presentation at IMC9<br />
(Hibbett et al. 2011), noted that the number of fungal OTUs<br />
added each year to GenBank that are based only on rDNA<br />
sequences (ENAS fungi) is now exceeding the number from<br />
fungi with cultures or specimens (Fig. 6). Ecologists face<br />
the prospect that most of the fungal species dwelling in their<br />
favourite environment can neither be cultivated nor collected;<br />
as a result they are going to have to rely on ENAS to assess<br />
the true fungal diversity. Each of these ecological studies may<br />
add hundreds or thousands of ENAS to GenBank. Already,<br />
searches of GenBank using a new ENAS mostly recover<br />
previously deposited ENASs, which are identified not by<br />
names but by numbers. Imagine two ecological studies, one<br />
where each new ENAS in tables or figures is associated with<br />
a numbered, existing ENAS and the other where the existing<br />
ENASs have been named – the reader would come away with<br />
ignorance on the one hand and understanding on the other.<br />
Fungal classification and nomenclature must respond to this<br />
challenge by developing a means of associating ENASs with<br />
names and the response must be timely.<br />
As discussed by Hibbett etal. (2011), fungi known only as<br />
ENAS can be named by comparison to named fungi already<br />
in GenBank. It seems important that this name be identified<br />
as attached to an ENAS rather than a culture or specimen,<br />
volume 2 · no. 2 117
John W. Taylor<br />
ARTICLE<br />
perhaps by appending ENAS as a suffix. Several essential<br />
<strong>issue</strong>s will have to addressed before ENAS naming can begin,<br />
among them the problems of sequencing errors, variation in<br />
rDNA sequence within an individual, and accommodation of<br />
all these new ENAS fungi in MycoBank (Hawksworth et al.<br />
2010). Perhaps most unsettlingly, the naming will have to be<br />
automated in some way because no one can possibly name<br />
the thousands of new sequences that will arise in each new<br />
environmental study.<br />
At this point, a reader might fairly ask, if separate<br />
“Deuteromycota” and “Eumycota” nomenclatural systems<br />
still remain separate 20 years after their merger became<br />
intellectually obvious, how could anyone possibly entertain<br />
thoughts about the acceptance of the automated description<br />
of fungi based only on DNA sequence? I see two steps<br />
to acceptance of ENAS fungi. The first step would be a<br />
published demonstration of the naming of ENAS fungi,<br />
echoing the aforementioned social activism already in play<br />
for One <strong>Fungus</strong> = One Name (Crous et al. 2006, Houbraken<br />
et al. 2010). The second step, acceptance of named ENAS<br />
fungi by the ICBN, is the tougher problem and is unlikely<br />
to occur quickly enough to satisfy the pressing needs of<br />
fungal ecologists. Here, social activism alone is not going to<br />
be sufficient largely due to the problem of organismal size,<br />
mentioned above, which is as old as Linnaeus. Mycologists<br />
cannot expect botanists to fully appreciate the problems<br />
created by working with microscopic organisms that can<br />
neither be routinely collected nor cultured. Mycology, to free<br />
itself from the legacy of botanical nomenclature, needs a<br />
nomenclatorial revolution.<br />
It is time for mycologists, who best understand the<br />
nomenclatorial needs peculiar to fungi, to design a<br />
nomenclatorial code for fungi. The timing could not be better<br />
because over the past two decades one of our own, David<br />
Hawksworth, has been helping to guide the development<br />
of the BioCode (Greuter et al. 2011, Hawksworth 2011).<br />
Modification of the draft BioCode to enable One <strong>Fungus</strong> =<br />
One Name and to accommodate ENAS fungi could produce<br />
a MycoCode that would be fully compatible with the BioCode.<br />
In considering microscopic organisms, a newly created<br />
MycoCode could also inspire those working on Bacteria,<br />
Archaea and other microscopic Eukarya. We mycologists<br />
have the need and, in the nomenclatorial committees of the<br />
International Mycological Association 2 and the Mycological<br />
Section of the International Union of Microbiological Societies,<br />
the means to accomplish this task. All that mycologists now<br />
lack is an excuse to do nothing.<br />
2<br />
Including the Nomenclature Committee for Fungi, which it is<br />
proposed be elected at International Mycological Congresses rather<br />
than at International Botanical Congresses as at present (Hawksworth<br />
et al. 2009, Norvell et al. 2010), and the International Commission on<br />
the Taxonomy of Fungi (a joint Commission with IUMS).<br />
ACKNOWLEDGEMENTS<br />
Thanks are due to Pedro Crous and Robert Samson of the CBS,<br />
Utrecht, who conceived of and hosted the One <strong>Fungus</strong> = One<br />
Name Conference, on 19–20 April 2011 in Amsterdam. JWT also<br />
acknowledges support from NSF DEB-0516511<br />
REFERENCES<br />
Barron GL (1968) The Genera of Hyphomycetes from Soil. Baltimore:<br />
Williams & Wilkins.<br />
Berbee ML, Taylor JW (1992) 18S ribosomal RNA gene sequence<br />
characters place the human pathogen Sporothrix schenckii in the<br />
genus Ophiostoma. Experimental Mycology 16: 87–91.<br />
Briquet J (1912) Règles internationales de al nomenclature botanique<br />
adoptées par le Congrès international de botanique de Vienne<br />
1905 dèuxieme édition mise au point d’après les décisions du<br />
congrès international de botanique de Bruxelles 1910. Jena:<br />
Commission de Rédaction du Congrès, Fischer Verlag.<br />
Bruns TD, Fogel R, White TJ, Palmer JD (1989) Accelerated<br />
evolution of a false-truffle from a mushroom ancestor. Nature<br />
339: 140–142.<br />
Bruns TD, White TJ, Taylor JW (1991) Fungal molecular systematics.<br />
Annual Review of Ecology and Systematics 22: 525–564.<br />
Burt A, Carter DA, Koenig GL, White TJ, Taylor JW (1996) Molecular<br />
markers reveal cryptic sex in the human pathogen Coccidioides<br />
immitis. Proceedings of the National Academy of Sciences, USA<br />
93: 770–773.<br />
Cole GT, Samson RA (1979) Patterns of Development in Conidial<br />
Fungi. London: Pitman.<br />
Crous PW, Slippers B, Wingfield MJ, Rheeder J, Marasas WFO,<br />
Philips AJL, Alves A, Burgess T, Barber P, Groenewald JZ (2006)<br />
Phylogenetic lineages in the Botryosphaeriaceae. Studies in<br />
Mycology 55: 235–253.<br />
Federal Register (2005) Possession, use and transfer of select<br />
agents and toxins; Final rule. 70. Federal Register 52 (18 March<br />
2005): 13294–13325 (13297).<br />
Fisher MC, KoenigGL, White TJ, Taylor JW (2002) Molecular and<br />
phenotypic description of Coccidioides posadasii sp. nov.,<br />
previously recognized as the non-California population of<br />
Coccidioides immitis. Mycologia 94: 73–84.<br />
Fuckel L (1870) Symbolae mycologicae. Beiträge zur Kenntnis der<br />
rheinischen Pilze. Jahrbücher des Nassauischen Vereins für<br />
Naturkunde 23-24: 1–495.<br />
Gottschalk M, Blanz PA (1984) Highly conserved 5S ribosomal RNA<br />
sequences in four rust fungi and atypical 5S rRNA secondary<br />
structure in Microstroma juglandis. Nucleic Acids Research 12:<br />
3951–3958.<br />
Greuter W, Garrity G, Hawksworth DL, Jahn R, Kirk PM, Knapp S,<br />
McNeill J, Michel E, Patterson DJ, Pyle R, Tindall B (2011) Draft<br />
BioCode (2011): principles and rules regulating the naming of<br />
organisms. Taxon 60: 201–212.<br />
Guadet J, Julien J, Lafay JF, Brygoo Y (1989) Phylogeny of some<br />
Fusarium species, as determined by large-subunit rRNA<br />
sequence comparison. Molecular Biology and Evolution 6: 227–<br />
242.<br />
118 <br />
ima fUNGUS
One <strong>Fungus</strong> = One Name: DNA and fungal nomenclature twenty years after PCR<br />
Gueho E, Kurtzman CP, Peterson SW (1989) Evolutionary affinities<br />
of heterobasidiomycetous yeast estimated from 18S and 25S<br />
ribosomal RNA seqeunce divergence. Systematic and Applied<br />
Microbiology 12: 230–236.<br />
Hawksworth DL (2011) Introducing the Draft BioCode (2011). Taxon<br />
60: 199–200.<br />
Hawksworth DL, Cooper JA, Crous PW, Hyde KD, Iturriaga T, Kirk<br />
PM, Lumbsch HT, May TW, Minter DW, Misra JK, Norvell L,<br />
Redhead SA, Rossman AY, Seifert KA, Stalpers JA, Taylor JW,<br />
Wingfield MJ (2010) Proposals to make the pre-publication<br />
deposit of key nomenclatural information in a recognized<br />
repository a requirement for valid publication of organisms<br />
treated as fungi under the Code. Mycotaxon 111: 514–519;<br />
Taxon 59: 660–662.<br />
Hawksworth DL, Crous PW, Dianese JC, Gryzenhout M, Norvell<br />
LL, Seifert KA (2009) Proposals to amend the Code to make it<br />
clear that it covers the nomenclature of fungi, and to modify the<br />
governance with respect to names of organisms treated as fungi.<br />
Taxon 58: 658–659; Mycotaxon 108: 1–4.<br />
Hawksworth DL, Crous PW, Redhead SA, Reynolds DR, Samson<br />
RA, Seifert KA, Taylor JW, Wingfield MJ, et al. (2011) The<br />
Amserdam declaration on fungal nomenclature. <strong>IMA</strong> <strong>Fungus</strong> 2:<br />
105–112.<br />
Hennig W (1966) Phylogenetic Systematics. Urbana: University of<br />
Illinois Press.<br />
Hibbett DS, Ohman A, Glotzer D, Nuhn M, Kirk P, Nilsson RH (2011)<br />
Progress in molecular and morphological taxon discovery in<br />
Fungi and options for formal classification of environmental<br />
sequences. Fungal Biology Reviews 25: 38–47.<br />
Houbraken J, Frisvad JC, Samson RA (2010) Taxonomy of Penicillium<br />
citrinum and related species. Fungal Diversity 44: 117-133.<br />
Hughes SJ (1953) Conidiophores, conidia, and classification.<br />
Canadian Journal of Botany 31: 577–659.<br />
Inderbitzin P, Shoemaker RA, O’Neill NR, Turgeon BG, Berbee ML<br />
(2006) Systematics and mating systems of two fungal pathogens<br />
of opium poppy: the heterothallic Crivellia papaveracea with a<br />
Brachycladium penicillatum asexual state and a homothallic<br />
species with a Brachycladium papaveris asexual state. Canadian<br />
Journal of Botany 84: 1304–1326.<br />
Jumpponen A, Jones KL (2009) Massively parallel 454 sequencing<br />
indicates hyperdiverse fungal communities in temperate Quercus<br />
macrocarpa phyllosphere. New Phytologist 184: 438–448.<br />
Jumpponen A, Jones KL (2010) Seasonally dynamic fungal<br />
communities in the Quercus macrocarpa phyllosphere differ<br />
between urban and nonurban environments. New Phytologist<br />
186: 496–513.<br />
Kendrick B (ed.)(1979) The Whole <strong>Fungus</strong>: the sexual-asexual<br />
synthesis. 2 vols. Ottawa: National Museums of Canada.<br />
Koufopanou V, Burt A, Taylor JW (1997) Concordance of gene<br />
genealogies reveals reproductive isolation in the pathogenic<br />
fungus Coccidioides immitis. Proceedings of the National<br />
Academy of Sciences, USA 94: 5478–5482.<br />
Kurtzman CP, Smiley MJ, Johnson CJ, Wickerham LJ, Fuson GB<br />
(1980) Two new and closely related heterothallic species,<br />
Pichia amyolphila and Pichia mississippiensis: characterization<br />
by hybridization and deoxyribonucleic acid reassociation.<br />
International Journal of Systematic Bacteriology 30: 208–216.<br />
Mason EW (1933) Annotated account of Fungi received at the<br />
Imperial Mycological Institute, List II (Fasc. 2). Mycological<br />
Papers 3: 1–67.<br />
Mason EW (1937) Annotated account of Fungi received at the<br />
Imperial Mycological Institute, List II, Fasc. 3 gen. part.<br />
Mycological Papers 4: 69–99.<br />
McNeill J, Barrie FR, Burdet HM, Demoulin V, Hawksworth DL,<br />
Marhold K, Nicolson DH, Prado J, Silva PC, Skog JE, Wiersema<br />
JH (eds) (2006) International Code of Botanical Nomenclature<br />
(Vienna Code) adopted by the Seventeenth Interational Botanical<br />
Congress, Vienna, Austria, July 2005. [Regnum Vegetabile No.<br />
146.] Ruggell: A.R.G. Gantner Verlag.<br />
Norvell LL, Hawksworth DL, Peterson RH, Redhead SA (2010) IMC9<br />
Edinburgh Nomenclature Sessions. <strong>IMA</strong> <strong>Fungus</strong> 1: 143–147;<br />
Mycotaxon 113: 503–511.<br />
Rabinow P (1996) Making PCR: a story of biotechnology. Chicago:<br />
University of Chicago Press.<br />
Raper KB, Fennell DI (1965) The Genus Aspergillus. Baltimore:<br />
Williams & Wilkins.<br />
Reynolds DR, Taylor JW (1991) DNA specimens and the ‘International<br />
code of botanical nomenclature’. Taxon 40: 311–315.<br />
Reynolds DR, Taylor JW (1992) Article 59: reinterpretation or<br />
revision? Taxon 42: 91–98.<br />
Reynolds DR, Taylor JW (eds) (1993) The Fungal Holomorph: mitotic,<br />
meiotic and pleomorphic speciation in fungal systematics.<br />
Wallingford: CAB International.<br />
Saccardo PA (1882) Sylloge Fungorum omnium hucusque<br />
cognitorum. Vol. 1. Pavia: PA Saccardo.<br />
Saiki RK, Gelfand DH, Stoffel S, Scharf SJ, Higuchi R, Horn GT, Mullis<br />
KB, Erlich H (1988) Primer-directed enzymatic amplification of<br />
DNA with a thermostable DNA polymerase. Science 239: 487–<br />
491.<br />
Taylor JW (1995) Making the Deuteromycota redundant: a practical<br />
integration of mitosporic and meiosporic fungi. Canadian Journal<br />
of Botany 73 (Suppl.): s754–s759.<br />
Taylor JW, Turner E, Townsend JP, Dettman JR, Jacobson D (2006)<br />
Eukaryotic microbes, species recognition and the geographic<br />
limits of species: examples from the kingdom Fungi. Philosophical<br />
Transactions of the Royal Society B-Biological Sciences 361:<br />
1947–1963.<br />
Tubaki K (1958) Studies on the Japanese Hyphomycetes, 5.<br />
Leaf and stem group with a discussion of the classification of<br />
Hyphomycetes and their perfect stages. Journal of the Hattori<br />
Botanical Laboratory 20: 142–244.<br />
Tulasne L-R, Tulasne C (1861) Selecta Fungroum Carpologia. Vol. 1.<br />
Paris: Imperial Typographer. [Reprint (1931) Oxford: Clarendon<br />
Press.]<br />
Vuillemin P (1910a) Les aleuriospores. Bulletin de la Société des<br />
Sciences du Nancy 12: 151–175.<br />
Vuillemin P (1910b) Les conidiospores. Bulletin de la Société des<br />
Sciences du Nancy 11: 129–172.<br />
Walker WF, Doolittle WF (1982) Redividing the basidiomycetes on<br />
the basis of 5S rRNA sequences. Nature 299: 723–724.<br />
Walker WF, Doolittle WF (1983) Systematics of basidiomycetes based<br />
on 5S rRNA sequences and other data. Nature 303: 731–732.<br />
Weresub LK, Pirozynski KA (1979) Pleomorphism of fungi as treated<br />
in the history of mycology and nomenclature. In: The Whole<br />
ARTICLE<br />
volume 2 · no. 2 119
John W. Taylor<br />
ARTICLE<br />
<strong>Fungus</strong> (B Kendrick, ed.) 1: 17–25. Ottawa: National Museums<br />
of Canada.<br />
White TJ, Bruns T, Lee S, Taylor W (1990) Amplification and direct<br />
sequencing of fungal ribosomal RNA genes for phylogenetics. In:<br />
PCR Protocols: a guide to methods and applications (M Innis et<br />
al., eds): 315–322. Academic Press, San Diego.<br />
120 <br />
ima fUNGUS
doi:10.5598/imafungus.2011.02.02.02<br />
<strong>IMA</strong> <strong>Fungus</strong> · volume 2 · no 2: 121–125<br />
Penicillium menonorum, a new species related to P. pimiteouiense<br />
Stephen W. Peterson 1 , Samantha S. Orchard 2 , and Suresh Menon 2<br />
1<br />
USDA, Agricultural Research Service, National Center for Agricultural Utilization Research, Bacterial Foodborne Pathogens and Mycology<br />
Research Unit, 1815 North University Street, Peoria, Illinois 61604 USA; corresponding author e-mail: Stephen.Peterson@ARS.USDA.GOV<br />
2<br />
Menon & Associates, Inc., P.O. Box 910033, San Diego, California 92191-0033 USA<br />
ARTICLE<br />
Abstract: Penicillium menonorum is described as a new monoverticillate, non-vesiculate species that resembles<br />
P. restrictum and P. pimiteouiense. On the basis of phylogenetic analysis of DNA sequences from four loci, P.<br />
menonorum occurs in a clade with P. pimiteouiense, P. vinaceum, P. guttulosum, P. rubidurum, and P. parvum.<br />
Genealogical concordance analysis was applied to P. pimiteouiense and P. parvum, substantiating the phenotypically<br />
defined species. The species P. rubidurum, P. guttulosum, and P. menonorum were on distinct branches statistically<br />
excluded from inclusion in other species and have distinct phenotypes.<br />
Key words:<br />
monoverticillate<br />
fungal systematics<br />
congruence analysis<br />
Penicillium<br />
Article info: Submitted: 13 April 2011; Accepted: 15 June 2011; Published: 29 September 2011.<br />
INTRODUCTION<br />
In the course of a screening program to find useful fungi for<br />
conversion of organic matter into high-value products such<br />
as lipid precursors to biofuels and animal feed formulations,<br />
a Penicillium isolated from garden soil in southern California<br />
was obtained that could not be placed with confidence<br />
in any described species. Sequencing of the ITS region<br />
was performed, with sequence analysis showing that this<br />
isolate is phylogenetically related to P. pimiteouiense. DNA<br />
distance from P. pimiteouiense suggested that it might be an<br />
undescribed species.<br />
Additional gene loci (β-tubulin, calmodulin, and DNA<br />
replication licensing factor Mcm7) were amplified and<br />
sequenced for this isolate and for phylogenetically and<br />
phenotypically similar species. On the basis of the phenotypic<br />
and phylogenetic distinctions, this isolate is described as a<br />
new species.<br />
MATERIALS AND METHODS<br />
Cultures (Table 1) may be obtained from the Agricultural<br />
Research Service Culture Collection (NRRL), Peoria, IL<br />
(http://nrrl.ncaur.usda.gov). The P. menonorum culture<br />
ex-type is available from the Agricultural Research Service<br />
Patent Culture Collection (http://nrrl.ncaur.usda.gov).<br />
Cultures were maintained on potato-dextrose agar (PDA)<br />
during the course of this study. Colony descriptions were<br />
based on 7 d growth of cultures on Czapek’s yeast autolysate<br />
agar (CYA), malt extract agar (MEA), and glycerol nitrate agar<br />
(G25N) at 25 °C, and on CYA at 5 °C and 37 °C as detailed<br />
by Pitt (1980). Some color names are taken from Ridgway<br />
(1912) and are designated with an upper case R and a plate<br />
number.<br />
Microscope slides were made by teasing apart bits<br />
of mycelium in a drop of lactic acid with cotton blue. A<br />
Zeiss axioscope with DIC optics was used for microscopic<br />
observations. Photomicrographs were taken with a Kodak<br />
14n digital camera attached to the microscope. Micro- and<br />
macro-photographs were sized and placed in a plate using<br />
Adobe Photoshop v. 6.0.1.<br />
Biomass for DNA extraction was grown in 125 mL flasks<br />
containing 25 mL malt extract (ME) broth incubated at 25<br />
°C on a rotary platform (200 rpm). Biomass ca. 0.5 g wet<br />
weight was collected by vacuum filtration, placed in micro<br />
centrifuge tubes, and freeze-dried. Freeze-dried mycelium<br />
was ground to a powder with a sterile pipette tip and DNA<br />
was extracted from the powdered biomass using the<br />
CTAB method. Purified DNA was stored in TE buffer (Tris<br />
10 mM, EDTA 1 mM, pH 8.0) at -20 °C until needed. DNA<br />
was amplified using the primers and conditions detailed<br />
in Peterson et al. (2010). Amplified DNA was prepared for<br />
sequencing using ExoSAP-IT (www.usbweb.com). DNA<br />
sequences were produced using DyeDeoxy v. 3.1 reagents<br />
and an ABI 3730 DNA sequencer (www.appliedbiosystems.<br />
com). Complementary strand sequences were assembled<br />
and corrected using Sequencher (www.genecodes.com).<br />
Finished sequences were aligned using CLUSTALW<br />
(Chenna et al. 2003), and maximum parsimony trees and<br />
bootstrap proportions were calculated using PAUP v. 4.0b10<br />
(Swofford 2003). MrBayes v. 3.12 (Huelsenbeck & Ronquist<br />
2001, Ronquist & Huelsenbeck 2003) was used to calculate<br />
Bayesian posterior probabilities. DNA sequences used in this<br />
© 2011 International Mycological Association<br />
You are free to share - to copy, distribute and transmit the work, under the following conditions:<br />
Attribution:<br />
You must attribute the work in the manner specified by the author or licensor (but not in any way that suggests that they endorse you or your use of the work).<br />
Non-commercial: You may not use this work for commercial purposes.<br />
No derivative works: You may not alter, transform, or build upon this work.<br />
For any reuse or distribution, you must make clear to others the license terms of this work, which can be found at http://creativecommons.org/licenses/by-nc-nd/3.0/legalcode. Any of the above conditions can be waived if you get<br />
permission from the copyright holder. Nothing in this license impairs or restricts the author’s moral rights.<br />
volume 2 · no. 2 121
Peterson, Orchard & Menon<br />
ARTICLE<br />
Table 1. Provenance of isolates used in this study.<br />
Species NRRL Accession No. Origin<br />
Penicillium erubescens MB335726 a (syn. Eupenicillium<br />
erubescens)<br />
6223 South Africa: Pretoria: isolated from nursery soil,<br />
1967, culture ex-type<br />
Penicillium guttulosum MB266689 907 USA: Utah: isolated from soil, 1927, culture ex-type<br />
Penicillium menonorum MB519297 50410 USA: California: isolated from garden soil, 2009,<br />
culture ex-type<br />
Penicillium parvum MB289101 (syn. Eupenicillium parvum) 2095<br />
Nicaragua: isolated from soil, July 1945, A.G.<br />
Kevorkian, culture ex-type<br />
6032 Papua-New Guinea: isolated from soil, ca. 1973, S.<br />
Udagawa, culture ex-type of P. papuanum MB319290<br />
35488 Ghana: Tafo: isolated from soil, ca. 1949<br />
35492 Venezuela: isolated from soil, ca. 1976, D.T. Wicklow<br />
Penicillium pimiteouiense MB460126 2063 New Guinea: isolated from tent cloth, ca. 1944, G.W.<br />
Martin<br />
25542 USA: Illinois: Peoria: isolated from human kidney cell<br />
culture plate, April 1996, J.T. Hjelle, culture ex-type<br />
26932 USA: Illinois: Peoria: isolated from human kidney cell<br />
culture plate, November 1997, M.A. Miller-Hjelle<br />
26933 USA: Illinois: Peoria: isolated from human kidney cell<br />
culture plate, November 1997, M.A. Miller-Hjelle<br />
28602 USA: Illinois: Peoria: isolated from human kidney cell<br />
culture plate, July 1998, J.T. Hjelle<br />
Penicillium rubidurum MB319295 (syn. Eupenicillium<br />
rubidurum)<br />
6033 Papua-New Guinea: isolated from soil, 1975, culture<br />
ex-type<br />
Penicillium vinaceum MB281754 739 USA: Utah: isolated from soil, 1927, culture ex-type<br />
740 Unknown: obtained from M.B. Morrow, 1936<br />
a<br />
MB=MycoBank (http://www.mycobank.org/).<br />
study are deposited in GenBank (www.ncbi.nlm.nih.gov)<br />
with accession numbers HQ646566–HQ646603, AF033460–<br />
AF033462, AF033464, AF037431, and AF037434. Data<br />
sets and tree diagrams are deposited at TREEBASE (www.<br />
treebase.org).<br />
The initial search to find phylogenetically related species<br />
was performed by BLAST searches of GenBank using the<br />
ITS sequence from the new species.<br />
Results<br />
Penicillium menonorum S.W. Peterson sp. nov.<br />
MycoBank MB519297<br />
(Fig. 1A–D)<br />
Etymology: Named for Menon & Associates whose scientists<br />
isolated the fungus.<br />
A speciebus aliis conidiophoris brevibus, conidiis scaberulis,<br />
colore in substrato nutritorio CYA pallide caesio atque augmento in<br />
temperatura 37 °C distinguendum.<br />
Typus: USA: California: isolated from garden soil, 2009 (BPI<br />
881018 – holotypus; culture ex-holotype NRRL 50410).<br />
Colonies on CYA (Fig. 1A) attaining 17–20 mm diam after 7 d<br />
growth at 25 °C, velutinous-silky, radially sulcate peripherally,<br />
centrally raised ca. 2–3 mm, sporulation moderate, central<br />
region pale bluish gray (court gray R-47), peripheral area white;<br />
no exudate or soluble pigments; no sclerotia or ascomata;<br />
reverse yellowish brown centrally (buckthorn brown R-15)<br />
to pale brownish-yellow (warm buff, R-15) peripherally. On<br />
MEA (Fig. 1B) attaining 17–19 mm diam after 7 d growth at<br />
25 °C, mycelium loosely woven, wooly, umbonate 3–4 mm<br />
deep centrally, sporulation moderate, white peripherally,<br />
court gray (R-47) centrally; no exudate or soluble pigments;<br />
no sclerotia or ascomata; reverse yellowish brown centrally<br />
to brownish yellow peripherally. On G25N attaining 8–10 mm<br />
diam after 7 d growth at 25 °C, umbonate, wooly 1–2 mm<br />
deep, white to court gray; no exudate or soluble pigment; no<br />
sclerotia or ascomata; reverse white to buff. Incubation for<br />
7 d on CYA at 5 °C produced no growth or germination of<br />
conidia. Incubation for 7 d on CYA at 37 °C produced colonies<br />
of 29–32 mm diam, resembling growth on CYA at 25 °C, but<br />
clear exudate moderately abundant, the reverse color is a<br />
darker, more uniform shade of brown. Conidiophores (Fig.<br />
1C) smooth-walled, hyaline, 5–15(–20) × 1.5–2.0 µm, nonvesiculate,<br />
with an apical whorl of (1–)2–5 phialides 5–7(–9)<br />
× 2.5–3.5 µm, conidia spherical to subspherical, (2–)2.5–3.5<br />
µm (Fig. 1D), with roughened to rugose surface.<br />
122 ima fUNGUS
Penicillium menonorum sp. nov.<br />
ARTICLE<br />
A B C D<br />
Fig. 1. Penicillium menonorum NRRL 50410. A. Colonies grown 7 d at 25 °C on CYA showing the radial sulcation and faint blue-gray central color<br />
characteristic of the species. Bar = 1 cm. B. Colonies grown 7 d at 25 °C on MEA having wooly consistency and darkened central area where the<br />
fungus is sporulating. Bar = 1 cm. C. Conidiophores, phialides and conidia. Bar = 10 µm. D. Roughened conidia. Bar = 10 µm.<br />
DNA sequences from the β-tubulin locus included all<br />
or part of 4 exon and 4 intron regions. After alignment the<br />
data set included 703 base positions. The calmodulin data<br />
included all or part of 4 exon and 3 intron regions and aligned<br />
with 726 base positions. The ID regions included the ITS1,<br />
ITS2, 5.8S rDNA, and ca. 650 bases from the 28S rDNA in<br />
an alignment of 1141 bases. DNA replication licensing protein<br />
(Mcm7) was composed of an amino acid coding region of 616<br />
bp length. Penicillium erubescens was chosen as the outgroup<br />
on the basis of phylogenetic trees previously published<br />
(Peterson et al. 1999, Peterson 2000).<br />
The most parsimonious trees, bootstrap proportion and<br />
Bayesian posterior probabilities for individual data sets<br />
were determined and the trees were compared for strongly<br />
supported contradictory branch points. Strongly supported<br />
nodes are those with > 90 % of the bootstrap sample and a<br />
Bayesian posterior probability of > 0.90. The individual locus<br />
trees contained no strongly supported contradictions that<br />
would preclude combining the data. The data from the four loci<br />
were combined to calculate a single phylogenetic tree (Fig. 2).<br />
The five isolates of P. pimiteouiense occur on a single<br />
strongly supported branch; three isolates of P. parvum<br />
and the single isolate of P. papuanum occur on a different<br />
strongly supported branch, and the two P. vinaceum isolates<br />
occur on another strongly supported branch. Penicillium<br />
rubidurum and P. guttulosum are most closely related to<br />
each other and form a sibling group to P. pimiteouiense,<br />
while P. menonorum is positioned basal in the tree to this<br />
three species branch.<br />
DISCUSSION<br />
Penicillium menonorum is similar phenotypically to P.<br />
pimiteouiense, P. restrictum, P. striatisporum, P. vinaceum,<br />
P. rubidurum, P. erubescens, and P. parvum. Penicillium<br />
restrictum, P. malacaense, P. kurssanovii, P. griseolum, and P.<br />
striatisporum, which phenotypically resemble P. menonorum,<br />
are phylogenetically positioned in different clades (Peterson<br />
& Horn 2009). Other species bearing some resemblance<br />
to P. menonorum are either not represented by extant extype<br />
cultures or the type cultures are not readily available.<br />
Penicillium menonorum differs from P. pimiteouiense by<br />
producing conidiophores in a basal layer rather than from<br />
aerial hyphae and a bluish gray (Court gray R-47) color<br />
on CYA versus white in P. pimiteouiense. Additionally, P.<br />
pimiteouiense produces yellow exudate and a brown soluble<br />
pigment, neither of which appear in P. menonorum after 7 d<br />
incubation. On different media (e.g., yeast extract malt agar<br />
incubated at 25 °C) or after extended incubation, a clear to<br />
rosy exudate often appears in P. menonorum. Penicillium<br />
restrictum produces somewhat longer conidiophores (up to<br />
60 µm) and has smaller colonies (< 10 mm diam) at 37 °C than<br />
P. menonorum (29–32 mm diam). Penicillium striatisporum<br />
produces rosy colored colonies on Czapek’s agar and has<br />
striate conidia. Penicillium vinaceum produces copious<br />
exudate in yellow to vinaceous colors, yellow to brown<br />
soluble pigments, and a dark brown colony reverse on CYA,<br />
and colonies grown at 37 °C are somewhat smaller (8–20<br />
mm diam) than those of P. menonorum. Penicillium parvum<br />
typically has mycelium that varies from white to yellow to red<br />
in color, while the P. menonorum mycelium is uniformly white.<br />
Penicillium parvum usually makes brown or purple-brown<br />
exudate, a brown soluble pigment, and has a colony reverse<br />
that is deep reddish-brown versus P. menonorum, which<br />
has no exudate or soluble pigments and a yellow brown<br />
colony reverse after 7 d incubation. Penicillium rubidurum<br />
produces white to orange or rosy-buff mycelium, red-brown<br />
exudate, a dark brown colony reverse, and produces conidia<br />
on M40Y medium but not on CYA. Penicillium menonorum<br />
produces no exudate or soluble pigment and has a yellow<br />
brown reverse and has abundant conidiogenesis on CYA.<br />
Penicillium erubescens produces white, pink or flesh color<br />
mycelium, reddish-brown exudate, and gray-red to magenta<br />
to vinaceous purple soluble pigments, with colony reverse<br />
either similarly colored or brown. Each of these species is<br />
volume 2 · no. 2<br />
123
Peterson, Orchard & Menon<br />
ARTICLE<br />
Combined data from BT2, CF, ID and Mcm7 loci, <br />
3186 total characters, 2798 are constant, 204 are <br />
variable not parsimony informaKve, 200 are parsimony <br />
InformaKve; 1 mpt, CI=0.8348, RC=0.7190 <br />
NRRL 2063 <br />
NRRL 28602 <br />
100/1.00 <br />
NRRL 26933 <br />
P. pimitiouiense<br />
64/0.94 <br />
NRRL 26932 <br />
81/1.00 <br />
NRRL 25542 T <br />
98/1.00 P. rubidurum NRRL 6033 <br />
P. gu+ulosum NRRL 907 <br />
P. menonorum NRRL 50410 T <br />
90/1.00 <br />
NRRL 35488 <br />
100/1.00 <br />
NRRL 35492 <br />
NRRL 2095 T <br />
P. papuanum NRRL 6032 T P. parvum <br />
100/1.00 <br />
P. vinaceum NRRL 739 T <br />
P. vinaceum NRRL 740 <br />
10 <br />
P. erubescens NRRL 6223 T <br />
Fig. 2. Phylogenetic tree calculated using maximum parsimony criterion for the concatenated data set composed of beta-tubulin, calmodulin, ITS<br />
and 28S rDNA, and DNA replication licensing protein (Mcm7). Bootstrap proportions/Bayesian posterior probabilities are placed on internodes.<br />
easily distinguished from P. menonorum on these bases.<br />
Raper & Thom (1949) regarded P. guttulosum to be a<br />
synonym of P. janthinellum, differing primarily by the production<br />
of copious amounts of exudate. Penicillium guttulosum as<br />
represented by Gilman & Abbott’s ex-type strain is distinct<br />
from P. janthinellum as well as the species studied here.<br />
Penicillium guttulosum cultures on CYA resemble the cultures<br />
of P. vinaceum, differing most noticeably in the production of<br />
dark purple exudate in large quantities, while P. vinaceum<br />
exudate is more red in color. Penicillium rubidurum colonies<br />
also resemble P. vinaceum and P. guttulosum but produce<br />
pale yellow exudate. Pitt (1980) treated P. papuanum as a<br />
124 ima fUNGUS
Penicillium menonorum sp. nov.<br />
synonym of P. parvum and they are in the same strongly<br />
supported clade (Fig. 2). Phenotypically, they are very similar<br />
to each other. Additional isolates of each species are needed<br />
to further assess the phylogenetic and phenotypic distinctions<br />
of these species.<br />
Phylogenetic systematics (Hennig 1966) is based on the<br />
principle that species must be monophyletic. Taylor et al. (2000)<br />
presented the genealogical concordance phylogenetic species<br />
recognition (GCPSR) concept as a means of determining the<br />
boundaries of species in fungi. Dettman et al. (2006) showed<br />
experimentally that GCPSR is effective in recognizing species<br />
boundaries in the genus Neurospora. GCPSR can be applied<br />
to P. pimiteouiense and P. parvum in this study and the species<br />
are supported by the GCPSR principles. Penicillium vinaceum,<br />
P. guttulosum, P. rubidurum, and P. menonorum are each on<br />
distinct branches, but the boundaries of the species cannot be<br />
determined from the single isolates available here. Phenotypic<br />
distinctions make each of these species recognizable and the<br />
phylogenetic placement of the species is consistent with the<br />
phenotypic descriptions of the species.<br />
Peterson SW, Jurjevic Z, Bills GF, Stchigel AM, Guarro J, Vega FE<br />
(2010) The genus Hamigera, six new species and multilocus<br />
DNA sequence based phylogeny. Mycologia 102: 847–864.<br />
Pitt JI (1980) ['1977'] The genus Penicillium and its teleomorphic<br />
states Eupenicillium and Talaromyces. Academic Press, UK.<br />
Raper KB, Thom C (1949) The genus Penicillium. Williams and<br />
Wilkins, USA.<br />
Ridgway R (1912) Color standards and color nomenclature.<br />
Published by the author, USA.<br />
Ronquist F, Huelsenbeck JP (2003) MrBayes3: Bayesian phylogenetic<br />
inference under mixed models. Bioinformatics 19: 1572–1574.<br />
Swofford DL (2003) PAUP*. Phylogenetic Analysis Using Parsimony<br />
(*and other methods). Version 4. Sinauer Associates, USA.<br />
Taylor JW, Jacobson DJ, Kroken S, Kasuga T, Geiser DM, Hibbett<br />
DS, Fisher MC (2000) Phylogenetic species recognition and<br />
species concepts in fungi. Fungal Genetics and Biology 31:<br />
21–32.<br />
ARTICLE<br />
ACKNOWLEDGEMENTS<br />
Amy McGovern provided highly skilled technical support that is<br />
greatly appreciated. Patricia Eckel kindly translated the diagnosis<br />
into Latin. Mention of trade names or commercial products in this<br />
publication is solely for the purpose of providing specific information<br />
and does not imply recommendation or endorsement by the U.S.<br />
Department of Agriculture. USDA is an equal opportunity provider<br />
and employer.<br />
REFERENCES<br />
Chenna R, Sugawara H, Koike T, Lopez R, Gibson TJ, Higgins DG,<br />
Thompson JD (2003) Multiple sequence alignment with the<br />
Clustal series of programs. Nucleic Acids Research 31: 3497–<br />
3500.<br />
Dettman JR, Jacobson DJ, Taylor JW (2006) Multilocus sequence<br />
data reveal extensive phylogenetic species diversity within the<br />
Neurospora discreta complex. Mycologia 98: 436–446.<br />
Hennig W (1966) Phylogenetic Systematics. (English Translation).<br />
Urbana: University of Illinois Press.<br />
Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference<br />
of phylogenetic trees. Bioinformatics 17: 754–755.<br />
Peterson SW, Corneli S, Hjelle JT, Miller-Hjelle MA, Nowak DM,<br />
Bonneau PA (1999) Penicillium pimiteouiense: a new species<br />
isolated from polycystic kidney cell cultures. Mycologia 91: 269–<br />
277.<br />
Peterson SW (2000) Phylogenetic analysis of Penicillium based on<br />
ITS and LSU-rDNA sequences. In: RA Samson & JI Pitt (eds),<br />
Classification of Penicillium and Aspergillus: Integration of<br />
modern taxonomic methods: 163–178. Harwood Publishers, UK.<br />
Peterson SW, Horn BW (2009) Penicillium parvulum and Penicillium<br />
georgiense, sp. nov. isolated from the conidial heads of<br />
Aspergillus species. Mycologia 101: 71–83.<br />
volume 2 · no. 2<br />
125
ARTICLE<br />
<br />
ima fUNGUS
doi:10.5598/imafungus.2011.02.02.03 <strong>IMA</strong> <strong>Fungus</strong> · volume 2 · no 2: 127–133<br />
What is Scirrhia?<br />
Pedro W. Crous 1 , Andrew M. Minnis 2 , Olinto L. Pereira 3 , Acelino C. Alfenas 3 , Rafael F. Alfenas 3 , Amy Y. Rossman 2 , and Johannes<br />
Z. Groenewald 1<br />
1<br />
CBS-KNAW Fungal Biodiversity Centre, Uppsalalaan 8, 3584 CT Utrecht, The Netherlands; corresponding author e-mail: p.crous@cbs.knaw.nl<br />
2<br />
Systematic Mycology & Microbiology Laboratory, USDA-ARS, Rm. 246, B010A, 10300 Baltimore Ave., Beltsville, MD 20705, USA<br />
3<br />
Departemento de Fitopatologia, Universidade Federal de Viçosa, 36.570 Viçosa, MG, Brazil<br />
ARTICLE<br />
Abstract: The ascomycetous genus Scirrhia is presently treated as a member of Dothideomycetidae, though<br />
uncertainty remains as to which family it belongs in Capnodiales, Ascomycota. Recent collections on stems of a<br />
fern, Pteridium aquilinum (Dennstaedtiaceae) in Brazil, led to the discovery of a new species of Scirrhia, described<br />
here as S. brasiliensis. Based on DNA sequence data of the nuclear ribosomal DNA (LSU), Scirrhia is revealed<br />
to represent a member of Dothideomycetes, Capnodiales, Mycosphaerellaceae. Scirrhia is the first confirmed<br />
genus in Mycosphaerellaceae to have well developed pseudoparaphyses and a prominent hypostroma in which<br />
ascomata are arranged in parallel rows. Given the extremely slow growth rate and difficulty in obtaining cultures<br />
of S. brasiliensis on various growth media, it appears that Scirrhia represents a genus of potentially obligate plant<br />
pathogens within Mycosphaerellaceae.<br />
Key words:<br />
Brazil<br />
Capnodiales<br />
Dothideomycetes<br />
ITS<br />
LSU<br />
Mycosphaerellaceae<br />
Pteridium<br />
systematics<br />
Article info: Submitted: 19 August 2011; Accepted: 25 September 2011; Published: 3 October 2011.<br />
Introduction<br />
The genus Scirrhia was originally known from four species<br />
(Fuckel 1870), and later lectotypified based on S. rimosa,<br />
a species known from stems of Phragmites australis<br />
(Poaceae) collected in Europe (Saccardo 1883). Based on<br />
its ascomata arranged in linear rows in a hypostroma of<br />
pseudoparenchymatal cells, bitunicate asci, hyaline, 1-septate<br />
ascospores, and the presence of pseudoparaphyses,<br />
Müller & von Arx (1962) placed the genus in Dothiorales,<br />
Mycosphaerellaceae, listing Scirrhodothis (Theissen &<br />
Sydow 1915), Scirrhophragma (Theissen & Sydow 1915)<br />
and Metameris (Theissen & Sydow 1915) as possible generic<br />
synonyms. All three genera have bitunicate asci, hyaline,<br />
1-septate ascospores, pseudoparaphyses, and stromata<br />
with immersed, longitudinally arranged ascomata, appearing<br />
to justify this opinion.<br />
In a later study, von Arx & Müller (1975) again placed<br />
Scirrhia in Dothideaceae, suborder Dothideineae in<br />
Dothideales. Barr (1972) questioned the synonymy<br />
of Scirrhodothis (lectotype species S. confluens),<br />
Scirrhophragma (type species S. regalis) and Metameris<br />
(type species M. japonica) under Scirrhia, and Sivanesan<br />
(1984) also chose not to list them as such, but included<br />
species that occurred on Gymospermae, namely S. acicola<br />
and S. pini. Barr (1987) chose to reinstate these generic<br />
synonyms, and placed Scirrhia together with Mycosphaerella<br />
in Dothideaceae, Dothideales. Although little is known about<br />
the three generic synonyms listed by Müller & von Arx<br />
(1962), the species on Gymnospermae are presently treated<br />
as separate genera, namely Lecanosticta (L. acicola) and<br />
Dothistroma (D. pini) (Crous et al. 2009a). Presently, the<br />
taxonomic position of Scirrhia remains obscure.<br />
During a recent visit to Brazil, we collected fresh material<br />
of a species of Scirrhia on stems of a fern. The aims of<br />
the present study are, therefore, to identify the species of<br />
Scirrhia, and to see if the taxonomic position of the genus<br />
could be resolved.<br />
Materials and methods<br />
Isolates<br />
Stems bearing ascomata were soaked in water for<br />
approximately 2 h, after which they were placed in the bottom<br />
of Petri dish lids, with the top half of the dish containing<br />
2 % malt extract agar (MEA), or potato-dextrose agar (PDA;<br />
Crous et al. 2009b). Ascospore germination patterns were<br />
examined after 24 h, and single ascospore cultures were<br />
established as described earlier (Crous et al. 1991, Crous<br />
1998). Colonies were subcultured onto PDA and oatmeal<br />
agar (OA) (Crous et al. 2009b) and incubated at 25 °C under<br />
continuous near-ultraviolet light to promote sporulation.<br />
Germinating ascospores died on MEA. Reference strains<br />
are maintained in the CBS-KNAW Fungal Biodiversity Centre<br />
(CBS), Utrecht, The Netherlands.<br />
© 2011 International Mycological Association<br />
You are free to share - to copy, distribute and transmit the work, under the following conditions:<br />
Attribution:<br />
You must attribute the work in the manner specified by the author or licensor (but not in any way that suggests that they endorse you or your use of the work).<br />
Non-commercial: You may not use this work for commercial purposes.<br />
No derivative works: You may not alter, transform, or build upon this work.<br />
For any reuse or distribution, you must make clear to others the license terms of this work, which can be found at http://creativecommons.org/licenses/by-nc-nd/3.0/legalcode. Any of the above conditions can be waived if you get<br />
permission from the copyright holder. Nothing in this license impairs or restricts the author’s moral rights.<br />
volume 2 · no. 2 127
Crous et al.<br />
ARTICLE<br />
Teratosphaeria fibrillosa GU214506<br />
Johansonia chapadiensis HQ423450<br />
98<br />
Zygophiala cryptogama FJ147157<br />
Zygophiala wisconsinensis FJ147158<br />
Schizothyrium pomi EF134948<br />
100 77 Schizothyrium pomi EF134949<br />
98 Mycosphaerella intermedia DQ246248<br />
71 Mycosphaerella marksii GQ852614<br />
92 Microcyclosporella mali GU570547<br />
Mycosphaerella madeirae DQ204756<br />
56<br />
57 Pseudocercosporella sp. FJ031991<br />
Mycosphaerella punctiformis EU167569<br />
Ramularia proteae EU707899<br />
20 changes<br />
88 Ramularia coleosporii GU214692<br />
Ramularia uredinicola GU214694<br />
Ramularia lamii JF700950<br />
Ramularia miae DQ885902<br />
94 54 Ramularia eucalypti JF700949<br />
Ramularia pratensis var. pratensis EU019284<br />
100<br />
Mycosphaerella endophytica DQ246255<br />
Mycosphaerella endophytica GQ852603<br />
62 Mycosphaerella pseudoendophytica DQ246253<br />
98<br />
Passalora daleae EU040236<br />
Mycosphaerella pini GQ852597<br />
60 Dothistroma pini GQ852596<br />
Passalora fulva DQ008163<br />
Legend to families:<br />
Pseudocercosporella bakeri GU570553<br />
79<br />
Incertae sedis<br />
72 Mycosphaerella microsora EU167599<br />
Schizothyriaceae<br />
57<br />
Passalora bellynckii GQ852618<br />
Passalora brachycarpa GQ852619<br />
Mycosphaerellaceae<br />
Mycosphaerella buckinghamiae EU707856<br />
62 Mycosphaerella africana DQ246258<br />
95 Mycosphaerella aurantia DQ246256<br />
Mycosphaerella ellipsoidea GQ852602<br />
100 Pseudocercospora vitis GU214483<br />
Pseudocercospora paraguayensis GQ852634<br />
66 59 Pseudocercospora natalensis DQ267576<br />
Cercosporella virgaureae GQ852585<br />
87 Ramulispora sorghi GQ852653<br />
Scirrhia brasiliensis CPC 18733<br />
53 Mycosphaerella podagrariae EU386700<br />
97 Mycosphaerella brassicicola EU167607<br />
Pseudocercosporella capsellae GU214662<br />
Septoria obesa GU214493<br />
82<br />
Mycosphaerella latebrosa GU214444<br />
Mycosphaerella berberidis EU167603<br />
54 Mycosphaerella harthensis EU167602<br />
Mycosphaerella flageoletiana EU167597<br />
74 Pseudocercosporella sp. GU214683<br />
Septoria apiicola GQ852674<br />
Mycosphaerella rubi EU167589<br />
Septoria convolvuli GQ852675<br />
Septoria senecionis GQ852678<br />
Septoria leucanthemi GQ852677<br />
Pseudocercosporella sp. GU214684<br />
Pseudocercosporella sp. GU214686<br />
Passalora dioscoreae GU214665<br />
Septoria lactucae GU214491<br />
68<br />
Cercospora janseana GU214405<br />
Cercospora sojina GU214655<br />
87 Cercospora zebrinae GU214657<br />
Pseudocercosporella sp. GU214685<br />
Pseudocercospora eucommiae GU214674<br />
Mycosphaerella linorum EU167590<br />
Septoria cucubali GU214698<br />
91<br />
Mycosphaerella coacervata EU167596<br />
Fig. 1. The first of 1000 equally most parsimonious trees obtained from a heuristic search with 100 random taxon additions of the LSU sequence<br />
alignment. The scale bar shows 10 changes, and bootstrap support values from 1000 replicates are shown at the nodes. The novel sequence<br />
generated for this study is shown in bold. Branches present in the strict consensus tree are thickened and the tree was rooted to a sequence of<br />
Teratosphaeria fibrillosa (GenBank accession GU214506).<br />
128 ima fUNGUS
What is Scirrhia?<br />
DNA isolation, amplification and analyses<br />
Genomic DNA was isolated from fungal mycelium grown on<br />
MEA, using the UltraCleanTM Microbial DNA Isolation Kit<br />
(MoBio Laboratories, Solana Beach, CA, USA) according<br />
to the manufacturer’s protocols. The primers V9G (de Hoog<br />
& Gerrits van den Ende 1998) and LR5 (Vilgalys & Hester<br />
1990) were used to amplify part of the nuclear rDNA operon<br />
spanning the 3’ end of the 18S rRNA gene (SSU), ITS 1, the<br />
5.8S rRNA gene, ITS 2 and the first 900 bases at the 5’ end<br />
of the 28S rRNA gene (LSU). The primers ITS4 (White et al.<br />
1990) and LSU1Fd (Crous et al. 2009c) were used as internal<br />
sequence primers to ensure good quality sequences over the<br />
entire length of the amplicon. The PCR conditions, sequence<br />
alignment, and subsequent phylogenetic analysis followed<br />
the methods of Crous et al. (2006, 2009d). Sequences were<br />
compared with the sequences available in NCBI’s GenBank<br />
nucleotide (nr) database using a megablast search and results<br />
are discussed in the relevant species notes where applicable.<br />
Based on the Blast results, the novel sequence was added to<br />
the alignment of Crous et al. 2010 (TreeBASE study S10980).<br />
Alignment gaps were treated as new character states.<br />
Sequences derived in this study were lodged at GenBank,<br />
the alignment in TreeBASE (www.treebase.org/treebase/<br />
index.html), and taxonomic novelties in MycoBank (www.<br />
MycoBank.org; Crous et al. 2004).<br />
Morphology<br />
Morphological descriptions are based on preparations made<br />
from host material in clear lactic acid, with 30 measurements<br />
determined per structure, and observations made with a Nikon<br />
SMZ1500 dissecting microscope, and with a Zeiss Axioscope<br />
2 microscope using differential interference contrast (DIC)<br />
illumination. Colony characters and pigment production were<br />
noted after 3 mo of growth on PDA and OA (Crous et al. 2009b)<br />
incubated at 25 ºC. Colony colours (surface and reverse) were<br />
rated according to the colour charts of Rayner (1970).<br />
RESULTS<br />
Phylogeny<br />
Approximately 1700 bases, spanning the ITS and LSU<br />
regions, were obtained from the sequenced culture. The<br />
LSU region was used in the phylogenetic analysis for the<br />
generic placement (Fig. 1) and ITS to determine speciesrank<br />
relationships (see notes under the species description).<br />
The manually adjusted LSU alignment contained 66 taxa<br />
including the outgroup sequence and, of the 749 characters<br />
used in the phylogenetic analysis, 104 were parsimonyinformative,<br />
51 were variable and parsimony-uninformative,<br />
and 594 were constant. Only the first 1000 equally most<br />
parsimonious trees were retained from the heuristic search,<br />
the first of which is shown in Fig. 1 (TL = 415, CI = 0.494, RI =<br />
0.851, RC = 0.421). The phylogenetic tree of the LSU region<br />
(Fig. 1) show that the obtained sequence clusters together<br />
with species belonging to Cercospora, Pseudocercospora,<br />
Pseudocercosporella and Septoria in Mycosphaerellaceae.<br />
Taxonomy<br />
Scirrhia rimosa (Alb. & Schwein.) Fuckel, Jb. nassau.<br />
Ver. Naturk. 23–24: 221 (1870).<br />
Basionym: Sphaeria rimosa Alb. & Schwein., Consp.<br />
Fung.: 13 (1805).<br />
(Fig. 2)<br />
Specimen examined: Czech Republic: Lipnik, on culms (stems) of<br />
Phragmites australis, June 1942 (Reliquiae Petrakianae Institut für<br />
Systematische Botanik Graz, exsiccati no. 1871, BPI 1111233).<br />
Hypostroma on upper leaf surfaces, dark brown to black,<br />
immersed beneath epidermis, becoming raised, irregularly<br />
ellipsoid, to 20 mm long, 5 mm wide. Ascomata immersed in<br />
brown stroma of textura angularis; arranged in parallel rows,<br />
opening by a 10–15 µm diam central ostiole; ascomata to<br />
250 µm diam, wall 10–20 µm wide. Asci 80–130 × 13–18<br />
µm, hyaline, smooth, cylindrical, sessile in fascicles, shortstipitate,<br />
bitunicate with ocular chamber, 3–4 µm diam.<br />
Pseudoparaphyses hyaline, cellular, smooth, distributed<br />
among asci, septate, branched, prominently constricted<br />
at septa, 5–8 µm diam, extending above asci. Ascospores<br />
hyaline, smooth, biseriate in asci, guttulate, fusoid-ellipsoidal,<br />
widest just above septum, tapering towards both ends, but<br />
more prominently towards lower end, prominently constricted<br />
at the median septum, thin-walled, (20–)21–23(–26) × 6–7(–<br />
7.5) µm; ascospores germinating on host with germ tubes at<br />
right angles to the long axis of spore, but remaining hyaline,<br />
not distorting.<br />
Notes: Despite several attempts, we have thus far been<br />
unable to recollect fresh material of this species on<br />
Phragmites, and thus no DNA sequence data are available.<br />
The nature of the hypostroma, ascomatal arrangement, asci,<br />
ascospores, and pseudoparaphyses, closely match that of<br />
the species described below. The specimen described and<br />
illustrated here agrees with the original description as well as<br />
locality and host of this species, occurring on stems. A later<br />
Schweinitz collection on Zizania (Poaceae) from the USA<br />
proved to be a misidentified Leptosphaeria zizaniicola (Ellis<br />
& Everhart 1892). We are unaware of any confirmed reports<br />
of Scirrhia rimosa from North America.<br />
Scirrhia brasiliensis Crous, O.L. Pereira, Alfenas &<br />
R.F. Alfenas, sp. nov.<br />
MycoBank MB560601<br />
(Fig. 3)<br />
Etymology: Named after the country where the holotype was<br />
collected, Brazil.<br />
Scirrhiae rimosae morphologice similis, sed asco saepe minus quam<br />
octosporo et ascosporis dissimiliter contractis.<br />
Typus: Brazil: Minas Gerais: Viçosa, Serra do Brigadeiro, S20°41.487,<br />
W42°29.875, on stems of Pteridium aquilinum (Dennstaedtiaceae),<br />
22 Aug. 2010, P.W. Crous, O.L. Pereira, A.C. Alfenas & R.F. Alfenas,<br />
ARTICLE<br />
volume 2 · no. 2<br />
129
Crous et al.<br />
ARTICLE<br />
Fig. 2. Scirrhia rimosa (BPI 1111233). A. Hypostroma on leaf of Phragmites australis. B. Horizontal section through ascomata. C–F. Asci<br />
intermingled among pseudoparaphyses. G. Ascus with basal stalk. H. Ascospores inside disintegrated ascus. Bars: A = 5 mm; B = 250 µm, all<br />
others = 10 µm.<br />
(CBS H-20538 – holotypus; cultures ex-holotype CPC 18734, 18735,<br />
18733 = CBS 128762). (GenBank accession numbers: CPC 18733,<br />
ITS JN197292, LSU JN197293).<br />
Stem cankers narrowly fusoid, brown, at times with redpurple<br />
margin, visible on shoots and stems of host, up to<br />
5 mm wide, 3 cm long; raised areas forming longitudinal<br />
cracks in the host epidermis through which erumpent<br />
ascomata protrude. Ascomata to 160 µm diam, 200 µm<br />
high, subepidermal, situated in rows in a brown hypostroma<br />
of textura angularis; wall consisting of to 6 layers of textura<br />
angularis, to 60 µm diam; cells brown, becoming flattened<br />
between ascomata, to 20 µm diam; ostioles ellipsoid, single,<br />
central, to 20 µm diam, situated on a dark brown, depressed<br />
neck, to 50 µm wide, 15 µm high; ostiole lined with hyaline,<br />
cylindrical periphyses with rounded ends, to 25 µm long, 3<br />
µm wide. Asci 90–130 × 8–11 µm, fasciculate, intermingled<br />
between pseudoparaphyses, subcylindrical, with thick wall,<br />
2–4 µm diam, inconspicuous apical chamber, 2–3 µm diam;<br />
wall appearing multi-layered with fissitunicate dehiscense,<br />
widest in middle of ascus, tapering to a long, thin, basal part<br />
and lobed base, to 9 µm wide; asci initially with 8 immature<br />
ascospores, of which some dissolve, leaving 4–6 mature<br />
ascospores per ascus. Pseudoparaphyses hyaline, cellular,<br />
smooth, septate, anastomosing, extending above asci, 3–5<br />
µm wide. Ascospores fusoid-ellipsoidal, hyaline, smooth,<br />
guttulate, obliquely uniseriate in asci, medianly 1-septate, at<br />
times constricted at the septum, widest in the middle of apical<br />
cell, tapering towards both ends, (17–)20–23(–27) × (5–)<br />
6–7(–8) µm; germinating from both ends, with germ tubes<br />
generally parallel to the long axis; spores neither darkening<br />
nor distorting.<br />
Culture characteristics: Colonies after 3 mo on PDA<br />
and OA with sparse aerial mycelium and even margins,<br />
olivaceous-grey (surface and reverse), reaching to 6 mm<br />
diam, extremely slow growing, with brown, diffuse pigment<br />
visible in agar surrounding colony; colonies sterile. Most<br />
ascospores died upon germination, and only a few of the<br />
germinated spores on PDA survived to form extremely slowgrowing<br />
colonies, suggesting this pathogen to be highly<br />
obligate in nature.<br />
Notes: The only culture of a species of Scirrhia presently<br />
available in the CBS culture collection, S. aspidiorum (CBS<br />
204.66, on Pteridium aquilinum, UK; GenBank accession<br />
number: ITS, JN197294) turned out to be related to Phoma<br />
herbarum (Didymellaceae, Pleosporales; de Gruyter et al.<br />
2010), and thus unrelated to S. brasiliensis. Although the<br />
specimen from which this culture was derived (ETH 2662)<br />
was not examined, examination of another collection under<br />
this name (The Netherlands, Baarn, Groenevelt, Pteridium<br />
130 ima fUNGUS
What is Scirrhia?<br />
ARTICLE<br />
Fig. 3. Scirrhia brasiliensis (ex-type CPC 18733). A. Fronds of Pteridium aquilinum. B, C. Stems with cankers extending into inner t<strong>issue</strong>. D.<br />
Hypostroma viewed from above. E–G, K. Sections through ascomata. H–J. Ostiolar area of ascomata, showing depressed neck. L–N. Bitunicate<br />
asci (arrow denotes apical chamber). O, P. Germinating ascospores. Q. Ascospores. Bars: B, C = 5 mm; D = 1 mm; E–H = 160 µm, all others<br />
= 10 µm.<br />
aquilinum, May 1962, J.A. von Arx, CBS H-17938), revealed<br />
a typical species of Scirrhia to be present on this fern, but no<br />
culture is presently available to confirm that it is the same<br />
taxon represented by CBS 204.66. Müller & von Arx (1962)<br />
treat S. aspidiorum in detail, citing ascospores as being 10–<br />
15 × 3–4 µm, thus being significantly smaller than those of<br />
S. brasiliensis. The ITS sequence of S. brasiliensis did not<br />
retrieve any highly identical hits with a megablast search of<br />
the NCBI’s GenBank nucleotide database; the closest hits<br />
represent species of Septoria, e.g. Septoria digitalis GenBank<br />
volume 2 · no. 2<br />
131
Crous et al.<br />
ARTICLE<br />
Fig. 4. Scirrhodothis confluens (isotype specimen CBS H-4806). A, B. Superficial view of stromata with imbedded ascomata. C. Asci. D. Broken<br />
asci with Didymella-like ascospores. Bars: A, B = 2 mm; C, D = 10 µm.<br />
FJ557246; Identities = 507/534 (95 %), Gaps = 8/534<br />
(1 %), and Cercospora, e.g. Cercospora piaropi GenBank<br />
HQ902254; Identities = 514/547 (94 %), Gaps = 9/547 (2 %)<br />
(data not shown).<br />
Discussion<br />
Persoon (1796) was the first to use the term “stroma” to<br />
define structures on or in which ascomata of the genus<br />
Sphaeria were borne. Fruisting (1867) introduced the terms<br />
“epistroma” for the hyaline pseudoparenchymatous crust<br />
formed in the outer layers of the host cortex, in which conidia<br />
were produced, and under which a “hypostroma” was formed<br />
in which ascomata developed. Based on the concepts of<br />
Fruisting, Ruhland (1900) introduced the terms “ectostroma”<br />
and “entostroma”. Blain (1927) subsequently defined Scirrhia<br />
rimosa as having a dothideoid epistroma and a hypostroma<br />
of compressed pseudoprosenchyma. The same hypostroma<br />
anatomy observed in S. rimosa was also found in S.<br />
aspidiorum and S. brasiliensis, and appears to be typical for<br />
the genus.<br />
Although the type species of Hadrotrichum, H. phragmitis,<br />
has been linked to S. rimosa as a possible anamorph of the<br />
genus Scirrhia (Fuckel 1870), this relationship has never<br />
been proven and has been disputed (von Arx & Müller 1975).<br />
The phylogenetic position of Hadrotrichum presently remains<br />
unknown, and this genus will have to be recollected to<br />
resolve this <strong>issue</strong>. Presently, no anamorph connections have<br />
been confirmed for Scirrhia, and none formed in culture for<br />
S. brasiliensis.<br />
Von Arx & Müller (1975) considered Scirrhodothis,<br />
Scirrhophragma, and Metameris to be synonyms of<br />
Scirrhia. Holm & Holm (1978) mentioned that although the<br />
taxa were closely related, the synonymy was controversial<br />
given different types of centrum development in some of the<br />
species. Barr (1992) regarded Scirrhia to represent a genus<br />
in Dothideales, while members of Metameris were found<br />
to have pseudoparaphyses and 1–2-septate, Didymellalike<br />
ascospores (Pleosporales). Furthermore, based on<br />
its small ascomata, broadly cylindrical to slightly obclavate<br />
asci with a short, thick, knob-like pedicels, as well as its<br />
monocotyledonous host preference, Zhang et al. (2012)<br />
suggested Metameris to belong to Phaeosphaeriaceae,<br />
though molecular data are still lacking to resolve its final<br />
placement.<br />
Lumbsch & Huhndorf (2010) list Scirrhia under<br />
Dothideaceae and Metameris under Phaeosphaeriaceae.<br />
The lectotype species of the genus Scirrhodothis, S. confluens<br />
(isotype CBS H-4806; Fig. 4), was examined in the present<br />
study and found to differ from Scirrhia in that the ascomata<br />
were locules formed in a stroma, not in linear rows in a<br />
hypostroma as in Scirrhia, and ascospores were Didymellalike.<br />
Although Scirrhodothis is clearly not a synonym of<br />
Scirrhia, the synonymy of these genera cannot be resolved<br />
in the absence of cultures, and they are best retained as<br />
separate entities until more data become available.<br />
Kirschner & Chen (2010) stated that the systematic<br />
position of Scirrhia was also in need of re-examination, and<br />
referred to a paper by Suetrong et al. (2009), who placed<br />
S. annulata (Juncaceae) within Mycosphaerellaceae.<br />
The clustering of S. brasiliensis in this family in the<br />
present study supports this finding and confirms<br />
Scirrhia as the first genus within Mycosphaerellaceae<br />
(Capnodiales, Dothideomycetes) with well developed<br />
pseudoparaphyses and an epi- and hypostroma. Other<br />
ascomycetous genera that have recently been shown to<br />
cluster in Mycosphaerellaceae include Phaeocryptopus<br />
and Rosenscheldiella (Winton et al. 2007, Sultan<br />
et al. 2011). Further collections are now required to<br />
resolve the questions: does S. rimosa cluster with<br />
S. brasiliensis, and what is the status of its reported<br />
synonyms, Scirrhodothis, Scirrhophragma and Metameris?<br />
132 ima fUNGUS
What is Scirrhia?<br />
Acknowledgements<br />
We thank the technical staff, Arien van Iperen (cultures), Marjan<br />
Vermaas (photo plates), and Mieke Starink-Willemse (DNA isolation,<br />
amplification and sequencing) for their invaluable assistance.<br />
References<br />
Arx von JA, Müller E (1975) A re-evaluation of the bitunicate<br />
ascomycetes with keys to families and genera. Studies in<br />
Mycology 9: 1–159.<br />
Barr ME (1972) Preliminary studies on the Dothideales in temperate<br />
North America. Contributions to the University of Michigan<br />
Herbarium 9: 523–638.<br />
Barr ME (1987) Prodomus to class Loculoascomycetes. Amherst,<br />
MA: M E Barr.<br />
Barr ME (1992) Additions to and notes on the Phaeosphaeriaceae<br />
(Pleosporales, Loculoascomycetes). Mycotaxon 43: 371–400.<br />
Blain WL (1927) Comparative morphology of dothideaceous and<br />
kindred stromata. Mycologia 19: 1–20.<br />
Crous PW (1998) Mycosphaerella spp. and their anamorphs<br />
associated with leaf spot diseases of Eucalyptus. Mycologia<br />
Memoir 21: 1–170.<br />
Crous PW, Barreto RW, Alfenas AC, Alfenas R, Groenewald JZ<br />
(2010) What is Johansonia? <strong>IMA</strong> <strong>Fungus</strong> 1: 117–122.<br />
Crous PW, Gams W, Stalpers JA, Robert V, Stegehuis G (2004)<br />
MycoBank: an online initiative to launch mycology into the 21st<br />
century. Studies in Mycology 50: 19–22.<br />
Crous PW, Groenewald JZ, Risède J-M, Simoneau P, Hyde KD (2006)<br />
Calonectria species and their Cylindrocladium anamorphs:<br />
species with clavate vesicles. Studies in Mycology 55: 213–226.<br />
Crous PW, Groenewald JZ, Summerell BA, Wingfield BD, Wingfield<br />
MJ (2009d) Co-occurring species of Teratosphaeria on<br />
Eucalyptus. Persoonia 22: 38–48.<br />
Crous PW, Schoch CL, Hyde KD, Wood AR, Gueidan C, de Hoog<br />
GS, Groenewald JZ (2009c) Phylogenetic lineages in the<br />
Capnodiales. Studies in Mycology 64: 17–47.<br />
Crous PW, Summerell BA, Carnegie AJ, Wingfield MJ, Hunter<br />
GC, Burgess TI, Andjic V, Barber PA, Groenewald JZ (2009a)<br />
Unravelling Mycosphaerella: do you believe in genera?<br />
Persoonia 23: 99–118.<br />
Crous PW, Verkley GJM, Groenewald JZ, Samson RA (eds) (2009b)<br />
Fungal Biodiversity. [CBS Laboratory Manual Series 1.] Utrecht:<br />
Centraalbureau voor Schimmelcultures.<br />
Crous PW, Wingfield MJ, Park RF (1991) Mycosphaerella nubilosa a<br />
synonym of M. molleriana. Mycological Research 95: 628–632.<br />
Ellis JB, Everhart BM (1892) The North American Pyrenomycetes.<br />
Published by authors, Newfield, NJ: Ellis & Everhart.<br />
Fruisting W (1867) Zur entwicklungsgeschichte der Pyrenomyceten.<br />
Botanisches Zentralblatt 25: 177–311.<br />
Fuckel L (1870) Symbolae mycologicae. Jahrbücher des<br />
Nassauischen Vereins für Naturkunde, Wiesbaden 23–24:<br />
1–459.<br />
Gruyter J de, Woudenberg JHC, Aveskamp MM, Verkley GJM,<br />
Groenewald JZ, Crous PW (2010) Systematic reappraisal of<br />
species in Phoma section Paraphoma, Pyrenochaeta and<br />
Pleurophoma. Mycologia 102: 1066–1081.<br />
Hoog GS de, Gerrits van den Ende AHG (1998) Molecular diagnostics<br />
of clinical strains of filamentous basidiomycetes. Mycoses 41:<br />
183–189.<br />
Holm L, Holm K (1978) Some pteridicolous ascomycetes. Botaniska<br />
Notiser 131: 97–115.<br />
Kirschner R, Chen C-J (2010) Periconiella species (anamorphic<br />
Mycosphaerellaceae) from Taiwan. Fungal Diversity 44: 135–<br />
148.<br />
Lumbsch HT, Huhndorf SM (2010) Myconet Volume 14. Part<br />
One. Outline of Ascomycota – 2009. Fieldiana, Life and Earth<br />
Sciences 1: 1–42.<br />
Müller E, von Arx JA (1962) Die Gattungen der didymosporen<br />
Pyrenomyceten. Beitrage zur Kryptogamenflora der Schweiz 11<br />
(2): 1–992.<br />
Persoon CH (1796) Observationes Mycologicae. Vol 1. Leipzig: PP<br />
Wolf.<br />
Rayner RW (1970) A Mycological Colour Chart. Kew: Commonwealth<br />
Mycological Institute.<br />
Ruhland W (1900) Untersuchungen zu einer morphologie der<br />
stromabildenen Sphaeriales. Hedwigia 39: 1–79.<br />
Saccardo PA (1883) Sylloge Fungorum. Vol. 2. Pyrenomycologiae<br />
Universae. Padua: P A Saccardo.<br />
Sivanesan A (1984) The Bitunicate Ascomycetes. Vaduz: J Cramer.<br />
Suetrong S, Schoch CL, Spatafora JW, Kohlmeyer J, Volkmann-<br />
Kohlmeyer B, Sakayaroj J, Phongpaichit S, Tanaka K, Hirayama<br />
K, Jones EBG (2009) Molecular systematics of the marine<br />
Dothideomycetes. Studies in Mycology 64: 155–173.<br />
Sultan, A, Johnston PR, Park D, Robertson AW (2011) Two new<br />
pathogenic ascomycetes in Guignardia and Rosenscheldiella<br />
on New Zealand’s pygmy mistletoes (Korthalsella: Viscaceae).<br />
Studies in Mycology 68: 237–247.<br />
Theissen F, Sydow H (1915) Die Dothideales. Kritisch-systematische<br />
Originaluntersuchungen. Annales Mycologici 13: 147–746.<br />
Vilgalys R, Hester M (1990) Rapid genetic identification and<br />
mapping of enzymatically amplified ribosomal DNA from several<br />
Cryptococcus species. Journal of Bacteriology 172: 4238–4246.<br />
White TJ, Bruns T, Lee J, Taylor J (1990) Amplification and direct<br />
sequencing of fungal ribosomal RNA genes for phylogenetics.<br />
In: Innis MA, Gelfand DH, Sninsky JJ, White TJ (eds), PCR<br />
Protocols: a guide to methods and applications: 315–322. San<br />
Diego: Academic Press.<br />
Winton LM, Stone JK, Hansen EM, Shoemaker RA (2007) The<br />
systematic position of Phaeocryptopus gaeumannii. Mycologia<br />
99: 240–252.<br />
Zhang Y, Crous PW, Schoch CL, Hyde KD (2012) Pleosporales.<br />
Fungal Diversity: in press.<br />
ARTICLE<br />
volume 2 · no. 2<br />
133
ARTICLE<br />
<br />
ima fUNGUS
doi:10.5598/imafungus.2011.02.02.04 <strong>IMA</strong> <strong>Fungus</strong> · volume 2 · no 2: 135–142<br />
A new species of Antherospora supports the systematic placement of its<br />
host plant<br />
Marcin Piątek 1 , Matthias Lutz 2 , Paul A. Smith 3 , and Arthur O. Chater 4<br />
ARTICLE<br />
1<br />
Department of Mycology, W. Szafer Institute of Botany, Polish Academy of Sciences, Lubicz 46, PL-31-512 Kraków, Poland; corresponding<br />
author e-mail: m.piatek@botany.pl<br />
2<br />
Organismische Botanik, Institut für Evolution und Ökologie, Universität Tübingen, Auf der Morgenstelle 1, D-72076 Tübingen, Germany<br />
3<br />
128 Llancayo Street, Bargoed, Mid Glamorgan, CF81 8TP, UK<br />
4<br />
Windover, Penyrangor, Aberystwyth, SY23 1BJ, UK<br />
Key words:<br />
Abstract: The morphology and phylogeny of anther smut specimens on Tractema verna collected in the United Molecular Analysis<br />
Kingdom were investigated using light microscopy, scanning electron microscopy and partial rDNA sequence Phylogeny<br />
analyses. The anther smut of Tractema verna shows similarity to Antherospora eucomis, A. scillae, A. tourneuxii, A. Plant Pathogens<br />
urgineae, A. vaillantii, and A. vindobonensis but differs in spore size range, spore wall thickness, host plant genera Scilla verna<br />
and considerable divergences of ITS and LSU sequences. Consequently, the smut is described here as a new Smut Fungi<br />
species, Antherospora tractemae. The host plant was formerly included in the genus Scilla (S. verna), but recently Tractema verna<br />
moved to a distinct genus Tractema. Molecular phylogenetic analyses reveal that Antherospora tractemae is sister Coevolution<br />
to the lineage of Muscari-parasitizing Antherospora and only distantly related to the Scilla-parasitizing Antherospora Ustilaginomycotina<br />
species. Thus, the phylogenetic placement of the smut fungus supports the systematic placement of its host plant.<br />
Article info: Submitted: 28 July 2011; Accepted: 30 September 2011; Published: 11 October 2011.<br />
INTRODUCTION<br />
The smut fungi sporulating in the anthers and on the<br />
surface of the inner floral organs of different Hyacinthaceae<br />
have recently been accommodated in a separate genus<br />
Antherospora (Bauer et al. 2008). Antherospora resides<br />
in the family Floromycetaceae (Urocystidales), together<br />
with the genus Floromyces, which produces sori in the<br />
inner floral organs of Anemarrhena asphodeloides<br />
(Agavaceae) (Vánky et al. 2008). Antherospora includes<br />
eight species, parasitic on hosts in seven different<br />
plant genera (Bauer et al. 2008, Vánky 2009). Despite<br />
phenotypic similarity, molecular phylogenetic analyses of<br />
Antherospora specimens parasitic on species of Muscari<br />
and Scilla have revealed significant genetic divergence<br />
between accessions from different host species (Bauer<br />
et al. 2008). For example, two closely related Central<br />
European Scilla species, S. bifolia and S. vindobonensis,<br />
harbour two morphologically similar but phylogenetically<br />
different Antherospora species. On the other hand, the<br />
phylogenetic results demonstrated that Antherospora<br />
vaillantii s. str. could infect two different hosts, Muscari<br />
comosum and M. neglectum (Bauer et al. 2008), indicating<br />
that some Antherospora spp. infect more than one host<br />
species. It is probable that host specificity is a widespread<br />
phenomenon and evolutionary driver in the genus<br />
Antherospora, similar, for example, to the anther smuts<br />
classified in the genus Microbotryum (Lutz et al. 2005,<br />
2008, Le Gac et al. 2007, Refrégier et al. 2008, Denchev<br />
et al. 2009, Kemler et al. 2009, Piątek et al. unpubl. data).<br />
Nevertheless, the DNA sequence data for Antherospora<br />
available in the NCBI’s GenBank nucleotide database<br />
is scant due, in part, to the inaccessibility of recently<br />
collected material. Much collecting and sequencing effort<br />
is necessary to understand the level of host specificity<br />
and the phylogenetic relationships within the genus.<br />
Infected specimens of Tractema verna were collected<br />
recently in Wales and the Outer Hebrides (United Kingdom).<br />
The host plant is commonly known by its synonym, Scilla<br />
verna, while its anther smut has been referred to as Ustilago<br />
vaillantii (Vánky 1994, Legon et al. 2005). This study aimed<br />
to clarify whether the collected specimens could be assigned<br />
to any of the described Antherospora species, especially to<br />
A. vaillantii or one of the recognized species sporulating in<br />
anthers of Scilla, or whether it represented a distinct species.<br />
A further aim was to check the phylogenetic affinity within<br />
the genus Antherospora and to expand the sampling of<br />
Antherospora species for which DNA sequence data are<br />
available.<br />
© 2011 International Mycological Association<br />
You are free to share - to copy, distribute and transmit the work, under the following conditions:<br />
Attribution:<br />
You must attribute the work in the manner specified by the author or licensor (but not in any way that suggests that they endorse you or your use of the work).<br />
Non-commercial: You may not use this work for commercial purposes.<br />
No derivative works: You may not alter, transform, or build upon this work.<br />
For any reuse or distribution, you must make clear to others the license terms of this work, which can be found at http://creativecommons.org/licenses/by-nc-nd/3.0/legalcode. Any of the above conditions can be waived if you get<br />
permission from the copyright holder. Nothing in this license impairs or restricts the author’s moral rights.<br />
volume 2 · no. 2 135
Piątek, Lutz, Smith & Chater<br />
ARTICLE<br />
MATERIALS AND METHODS<br />
Specimen sampling and documentation<br />
The specimens examined during the present work are listed in<br />
Table 1. The voucher specimens are deposited in KR, KRAM<br />
F and H.U.V. The latter abbreviation refers to the personal<br />
collection of Kálmán Vánky named as Herbarium Ustilaginales<br />
Vánky (Gabriel-Biel-Str. 5, D-72076 Tübingen, Germany). The<br />
nomenclatural novelty was registered in MycoBank (www.<br />
MycoBank.org, Crous et al. 2004).<br />
Nomenclature of anther smuts on<br />
Hyacinthaceae<br />
The nomenclature of anther smuts on Hyacinthaceae follows<br />
Bauer et al. (2008) and Vánky (2009). The collective name<br />
Ustilago vaillantii (syn. Vankya vaillantii) refers to all anther<br />
smuts on hyacinthaceous genera and species. The name<br />
Antherospora vaillantii refers to a species complex on<br />
Muscari spp., while Antherospora vaillantii s. str. refers to the<br />
species in its narrow sense (Bauer et al. 2008).<br />
Morphological examination<br />
Dried fungal teliospores of the investigated specimens were<br />
mounted in lactic acid, heated to boiling point and cooled, and<br />
then examined under a Nikon Eclipse 80i light microscope at a<br />
magnification of ×1000, using Nomarski optics (DIC). Spores<br />
were measured using NIS-Elements BR 3.0 imaging software.<br />
Spore size range, and the mean and standard deviation of<br />
the size of 50 measured spores were calculated for each<br />
investigated specimen (Table 1). The species description<br />
includes the combined values from all measured specimens.<br />
LM micrographs were taken with a Nikon DS-Fi1 camera.The<br />
ornamentation of the spore surface was studied using scanning<br />
electron microscopy (SEM). For this purpose, dry spores were<br />
mounted on carbon tabs and fixed to an aluminium stub with<br />
double-sided transparent tape. The tabs were sputter-coated<br />
with carbon using a Cressington sputter-coater and viewed<br />
with a Hitachi S-4700 scanning electron microscope, with a<br />
working distance of ca. 12−13 mm. SEM micrographs were<br />
taken in the Laboratory of Field Emission Scanning Electron<br />
Microscopy and Microanalysis at the Institute of Geological<br />
Sciences, Jagiellonian University, Kraków (Poland).<br />
Table 1. List of specimens, with host plants, GenBank accession numbers, spore size range, mean spore sizes with standard deviation, and<br />
reference specimens, newly examined in the course of this study.<br />
Smut species Host species GenBank<br />
acc. no. (ITS/<br />
LSU)<br />
Spore size<br />
range (µm)<br />
Average spore<br />
size with standard<br />
deviation (µm)<br />
Reference specimens 1<br />
Antherospora<br />
tractemae<br />
Antherospora<br />
tractemae<br />
Antherospora<br />
tractemae<br />
Antherospora<br />
tractemae<br />
Antherospora<br />
tractemae<br />
Floromyces<br />
anemarrhenae<br />
Tractema verna<br />
Tractema verna<br />
Tractema verna<br />
Tractema verna<br />
Tractema verna<br />
Anemarrhena<br />
asphodeloides<br />
JN104589/<br />
JN104590<br />
JN204283/<br />
JN204279<br />
JN204284/<br />
JN204280<br />
JN204285/<br />
JN204281<br />
JN204286/<br />
JN204282<br />
7.0–12.0 ×<br />
6.5–9.5<br />
7.5–14.5(–16.5)<br />
× 7.0–9.5(–10.5)<br />
6.5–12.5(–16.0)<br />
× (5.5–)6.0–<br />
9.5(–10.0)<br />
7.0–11.5(–15.0)<br />
× 6.0–10.5<br />
(7.0–)8.0–<br />
13.0(–14.0) ×<br />
6.0–10.5(–11.5)<br />
9.3 ± 1.3 × 7.8 ± 0.7 UK, Scotland, Outer Hebrides,<br />
Sgeir Ghlas Leac an Aiseig, Lewis,<br />
NA–993–215 [grid reference on<br />
UK national grid], 6 May 2010, P.A.<br />
Smith, KR 28182<br />
10.5 ± 2.1 × 8.5<br />
± 0.8<br />
UK, Wales, Ceredigion, Llangranog<br />
Head, SN–312–551 [grid reference<br />
on UK national grid], 19 April 2011,<br />
A.O. Chater, KRAM F-48879 –<br />
holotype<br />
9.3 ± 1.8 × 7.7 ± 0.9 UK, Wales, Ceredigion, 100 m SW<br />
of mouth of Cwm Soden, SN–361–<br />
582 [grid reference on UK national<br />
grid], 29 April 2011, A.O. Chater,<br />
KRAM F-48878<br />
9.6 ± 1.7 × 8.2 ± 1.1 UK, Wales, Ceredigion, 200 m NE<br />
of Mwnt church, SN–196–521 [grid<br />
reference on UK national grid], 6<br />
May 2011, A.O. Chater, KRAM<br />
F-48877<br />
10.1 ± 1.6 × 8.6<br />
± 1.2<br />
UK, Wales, Ceredigion, 500 m E<br />
of Mwnt church, SN–200–521 [grid<br />
reference on UK national grid], 6<br />
May 2011, A.O. Chater, KRAM<br />
F-48876<br />
JN104591/- not analysed not analysed China, Inner Mongolia, Chifeng<br />
city (Ulanhad), Hongshan Distr.,<br />
Hongshan, 15 July 2007, T.Z. Liu,<br />
H.U.V. 21482<br />
1<br />
H.U.V. – Herbarium Ustilaginales Vánky, Gabriel-Biel-Str. 5, D-72076 Tübingen, Germany; KR – Herbarium of the Staatliches Museum<br />
für Naturkunde, Karlsruhe, Germany; KRAM F – Mycological Herbarium of the W. Szafer Institute of Botany, Polish Academy of Sciences,<br />
Kraków, Poland.<br />
136 ima fUNGUS
Antherospora tractemae sp. nov.<br />
A. vindobonensis EF653990/EF653972 on S. vindobonensis<br />
A. vindobonensis EF653995/EF653977 on S. vindobonensis<br />
A. vindobonensis EF653989/EF653971 on S. vindobonensis<br />
85/97/94<br />
A. vindobonensis EF653993/EF653975 on S. vindobonensis<br />
ssp. borhidiana<br />
A. vindobonensis EF653994/EF653976 on S. vindobonensis<br />
ARTICLE<br />
100/100/99<br />
A. vindobonensis EF653991/EF653973 on S. vindobonensis<br />
A. scillae EF653983/EF653965 on S. bifolia<br />
87/100/91<br />
A. scillae EF653985/EF653967 on S. bifolia<br />
84/70/93<br />
A. scillae EF653984/EF653966 on S. bifolia<br />
100/100/100<br />
100/100/100<br />
77/99/96<br />
87/100/68<br />
91/100/93<br />
70/59/80<br />
81/-/-<br />
81/97/83<br />
A. scillae EF653992/EF653974 on S. bifolia<br />
A. scillae EF653996/EF653978 on S. bifolia<br />
A. vaillantii EF653986/EF653968 on M. tenuiflorum<br />
A. vaillantii EF653988/EF653970 on M. tenuiflorum<br />
A. vaillantii EF653987/EF653969 on M. tenuiflorum<br />
A. vaillantii EF653982/EF653964 on M. neglectum<br />
A. vaillantii EF653997/EF653979 on M. comosum<br />
A. vaillantii EF653998/EF653980 on M. neglectum<br />
A. tractemae JN104589/JN104590 on T. verna<br />
Floromycetaceae<br />
A. tractemae JN204283/JN204279 on T. verna<br />
99/100/98<br />
A. tractemae JN204284/JN204280 on T. verna<br />
A. tractemae JN204285/JN204281 on T. verna<br />
A. tractemae JN204286/JN204282 on T. verna<br />
Floromyces anemarrhenae JN104591/EU221284<br />
76/-/-<br />
Melanustilospora ari EF635904/EF517924<br />
-/59/73 Vankya ornithogali EF635910/EF210712<br />
-/-/56<br />
Ustacystis waldsteiniae DQ875356/AF009880<br />
60/100/99<br />
Melanoxa oxalidis EF635907/EF635908<br />
95/100/84<br />
Urocystis colchici DQ875355/DQ838576<br />
Flamingomyces ruppiae EF635909/DQ185436<br />
Mundkurella kalopanacis DQ875351/AF009869<br />
0.005 substitutions/site<br />
Fig. 1. Hypothesis on phylogenetic relationships within the sampled Urocystidales based on neighbour-joining analysis of an alignment of<br />
concatenated ITS + LSU base sequences using the TrN + G model of DNA substitution. The topology was rooted with the urocystidacean<br />
species. NJ bootstrap values of 1000 replicates are indicated before slashes, numbers on branches between slashes are estimates for a<br />
posteriori probabilities, numbers on branches after slashes are ML bootstrap support values. A. = Antherospora, M. = Muscari, S. = Scilla, T. =<br />
Tractema.<br />
Urocystidaceae<br />
DNA extraction, PCR, and sequencing<br />
Genomic DNA was isolated directly from the herbarium<br />
specimens. For methods of isolation and crushing of fungal<br />
material, DNA extraction, amplification, purification of PCR<br />
products, sequencing, and processing of the raw data see<br />
Lutz et al. (2004). ITS 1 and ITS 2 regions of the rDNA<br />
including the 5.8S rDNA (ITS) were amplified using the<br />
primer pair ITS1-F (Gardes & Bruns 1993) and ITS4 (White et<br />
al. 1990). The 5´-end of the nuclear large subunit ribosomal<br />
DNA (LSU) was amplified using the primer pairs LR0R and<br />
LR5 or NL1 and NL4, respectively (O´Donnell 1992, 1993,<br />
White et al. 1990). Primers were used for both PCR and cycle<br />
sequencing. For amplification the annealing temperature was<br />
adjusted to 45 °C. DNA sequences determined in this study<br />
were deposited in GenBank. GenBank accession numbers<br />
are given in Fig. 1 and Table 1.<br />
Phylogenetic analyses<br />
In addition to the sequences of Antherospora sp. on Tractema<br />
verna (ITS and LSU) and Floromyces anemarrhenae (ITS)<br />
newly obtained in this study, sequences from GenBank of<br />
the following species were used for molecular phylogenetic<br />
analyses (Begerow et al. 1997, 2006, Bauer et al. 2007,<br />
2008, Vánky et al. 2008, Lutz et al. in press): Antherospora<br />
volume 2 · no. 2<br />
137
Piątek, Lutz, Smith & Chater<br />
ARTICLE<br />
Table 2. ITS and LSU sequence divergences of Antherospora species used in phylogenetic analyses.<br />
Smut species Antherospora scillae Antherospora vaillantii Antherospora<br />
vindobonensis<br />
Host Scilla bifolia Muscari<br />
comosum<br />
Muscari<br />
neglectum<br />
Muscari<br />
tenuiflorum<br />
Scilla<br />
vindobonensis<br />
Antherospora tractemae ITS (653 a ) 3.4–3.5 % (22–23 b ) 1.8–2.0 % 1.4–2.0 % (9–13) 5.7–6.0 % (37–39) 3.5–3.7 % (23–24)<br />
on Tractema verna<br />
(no. of characters) LSU (658) 1.5–1.7 % (10–11)<br />
(12–13)<br />
0.9 % (6) 0.9–1.2 % (6–8) 1.2–1.4 % (8–9) 1.7–1.8 % (11–12)<br />
a<br />
A total number of nucleotide characters.<br />
b<br />
The number of different nucleotide characters.<br />
scillae, A. vaillantii, A. vindobonensis, Flamingomyces<br />
ruppiae, Floromyces anemarrhenae, Melanoxa oxalidis,<br />
Melanustilospora ari, Mundkurella kalopanacis, Urocystis<br />
colchici, Ustacystis waldsteiniae, and Vankya ornithogali<br />
(GenBank accession numbers included in Fig. 1).<br />
To elucidate the phylogenetic position of the Antherospora<br />
specimens from Tractema verna their concatenated ITS +<br />
LSU sequences were analysed within a dataset covering all<br />
sequences of Floromycetaceae available in GenBank and<br />
representatives of all genera of Urocystidaceae. If present in<br />
GenBank, the respective type species were used.<br />
Sequence alignment was obtained using MAFFT v. 6.853<br />
(Katoh et al. 2002, 2005, Katoh & Toh 2008) using the L-INS-i<br />
option. To obtain reproducible results, manipulation of the<br />
alignment by hand as well as manual exclusion of ambiguous<br />
sites were avoided as suggested by Giribet & Wheeler (1999)<br />
and Gatesy et al. (1993), respectively. Highly divergent<br />
portions of the alignment were omitted using GBlocks 0.91b<br />
(Castresana 2000) with the following options: “Minimum<br />
Number of Sequences for a Conserved Position” to 16,<br />
“Minimum Number of Sequences for a Flank Position” to 16,<br />
“Maximum Number of Contiguous Non-conserved Positions”<br />
to 8, “Minimum Length of a Block” to 5 and “Allowed Gap<br />
Positions” to “With half”.<br />
The resulting alignment [new number of positions: 1308<br />
(65 % of the original 1990 positions) number of variable<br />
sites: 300] was used for phylogenetic analyses using<br />
Neighbour-Joining (NJ), a Bayesian Approach (BA) and<br />
Maximum Likelihood (ML). For NJ analysis the data were<br />
first analysed with Modeltest 3.7 (Posada & Crandall 1998)<br />
to find the most appropriate model of DNA substitution. The<br />
hierarchical likelihood ratio test proposed the TrN + G DNA<br />
substitution model. Bootstrap values were calculated from<br />
1000 replicates. NJ analyses were carried out using PAUP<br />
v. 4.0b10 (Swofford 2001). For BA a Bayesian approach to<br />
phylogenetic inference using a Markov chain Monte Carlo<br />
technique was used as implemented in the computer program<br />
MrBayes v. 3.1.2 (Huelsenbeck & Ronquist 2001, Ronquist &<br />
Huelsenbeck 2003). Four incrementally heated simultaneous<br />
Markov chains were run over 5 000 000 generations using<br />
the general time reversible model of DNA substitution with<br />
gamma distributed substitution rates and estimation of<br />
invariant sites, random starting trees and default starting<br />
parameters of the DNA substitution model as recommended<br />
by Huelsenbeck & Rannala (2004). Trees were sampled<br />
every 100th generation, resulting in an overall sampling of<br />
50 001 trees. From these, the first 5 001 trees were discarded<br />
(burnin = 5 001). The trees sampled after the process had<br />
reached stationarity (45 000 trees) were used to compute a<br />
50 % majority rule consensus tree to obtain estimates for the<br />
a posteriori probabilities of groups of species. This Bayesian<br />
approach to phylogenetic analysis was repeated five times to<br />
test the independence of the results from topological priors<br />
(Huelsenbeck et al. 2002).<br />
ML analysis (Felsenstein 1981) was conducted with the<br />
RAxML v. 7.2.6 software (Stamatakis 2006), using raxmlGUI<br />
(Silvestro & Michalak 2010), invoking the GTRCAT and the<br />
rapid bootstrap option (Stamatakis et al. 2008) with 1000<br />
replicates.<br />
In line with Vánky et al. (2008), trees were rooted with the<br />
urocystidacean species Flamingomyces ruppiae, Melanoxa<br />
oxalidis, Melanustilospora ari, Mundkurella kalopanacis,<br />
Urocystis colchici, Ustacystis waldsteiniae, and Vankya<br />
ornithogali.<br />
RESULTS<br />
Morphological analyses<br />
The examined specimens on Tractema verna produced<br />
olivaceous sori with teliospores in all anthers of the<br />
inflorescences. The spores in all specimens were verruculose,<br />
variable in shape and size within particular collections, and<br />
variable in spore size range and mean spore size between<br />
different collections (Table 1). The spore wall was twolayered,<br />
although the layers were not always clearly visible<br />
in some spores. The detailed morphological characteristics<br />
of anther smut on Tractema verna are included in the species<br />
description and depicted in Fig. 2.<br />
Phylogenetic analyses<br />
The ITS sequences of the five Tractema verna anther smut<br />
specimens analysed differed in one base pair (0.15 %) from<br />
each other or were identical, LSU sequences were identical.<br />
ITS and LSU sequence divergences of Antherospora species<br />
used in phylogenetic analyses are included in Table 2.<br />
The different runs of BA that were performed and the ML<br />
analyses yielded consistent topologies which were congruent<br />
138 ima fUNGUS
Antherospora tractemae sp. nov.<br />
to the results of the NJ analysis in respect to well supported<br />
branchings (a posteriori probability greater than 54, ML<br />
bootstrap support values greater than 29). To illustrate the<br />
results, the phylogenetic hypothesis resulting from the NJ<br />
analysis is presented in Fig. 1. Bootstrap values from the NJ<br />
analysis are indicated on branches before slashes, estimates<br />
for a posteriori probabilities are indicated between slashes,<br />
numbers on branches after slashes are ML bootstrap support<br />
values.<br />
In all analyses the Antherospora species included in<br />
previous work (Bauer et al. 2008) were inferred with high<br />
support values, and phylogenetic relationships between<br />
floromycetacean species were as in Bauer et al. (2008)<br />
and Vánky et al. (2008). The Antherospora specimens from<br />
Tractema verna formed a well supported clade that clustered<br />
as a sister group of Antherospora vaillantii with high (NJ, BA)<br />
to moderate (ML) support values. Thus, the Antherospora<br />
specimens from Tractema verna were well separated from<br />
the Antherospora species growing on Scilla species, A.<br />
scillae and A. vindobonensis.<br />
on Tractema verna, 6 May 2011, A.O. Chater (KRAM F-48877);<br />
Ceredigion, 500 m E of Mwnt church, on Tractema verna, 6 May<br />
2011, A.O. Chater (KRAM F-48876).<br />
Ecology: The infected plants were found in April and May,<br />
peak flowering time for Tractema verna. The habitats for all<br />
the collections are broadly similar, consisting of short coastal<br />
turf on shallow soils. The Outer Hebrides locality is on pockets<br />
of maritime peat on a rocky substrate, referable to the MC10<br />
Festuca rubra–Plantago spp. maritime grassland community<br />
(plant community nomenclature follows Rodwell et al. 1991–<br />
2000). The Welsh sites are in coastal heath vegetation<br />
referable to the H7 Calluna vulgaris–Scilla verna heath and<br />
H8d Scilla verna subcommunity of Calluna vulgaris–Ulex<br />
gallii heath, as well as in the Armeria subcommunity of the<br />
MC10 community. Tractema verna is locally common around<br />
the rocky western coasts of Britain, and occurs in a number of<br />
maritime communities. The smut often infects large numbers<br />
of individuals within a population, and its incidence varies<br />
greatly from year to year.<br />
ARTICLE<br />
Taxonomy<br />
Antherospora tractemae M. Piątek & M. Lutz, sp. nov.<br />
MycoBank MB563318<br />
(Fig. 2)<br />
Etymology: Named after the host plant genus.<br />
Sori in antheris Tractemae vernae. Massa sporarum pulverulenta,<br />
olivaceo-brunnea. Sporae globosae, subglobosae, late ellipsoideae,<br />
late ovales, nonnumquam elongatae, pyriformae vel asymmetricae,<br />
6.5–14.5(–16.5) × (5.5–)6.0–10.5(–11.5) µm, olivaceae vel flavidobrunneae,<br />
parietibus 0.5–1.3 µm crassis, dense verruculosis, a<br />
latere visae fere levigatae vel subtiliter sinuatae.<br />
Typus: UK: Wales: Ceredigion, Llangranog Head, on Tractema verna<br />
(syn. Scilla verna), 19 Apr. 2011, A.O. Chater (KRAM F-48879 –<br />
holotypus; ITS/LSU sequences GenBank accession nos JN204283<br />
and JN204279).<br />
Parasitic on Tractema verna. Sori in the anthers, producing<br />
olive-brown, powdery mass of spores inside the pollen sacs.<br />
Infection systemic, all anthers of a plant infected. Spores<br />
globose, subglobose, broadly ellipsoidal, broadly ovoid,<br />
sometimes elongated, pyriform or asymmetrical, 6.5–14.5(–<br />
16.5) × (5.5–)6.0–10.5(–11.5) µm [av. ± SD, 9.8 ± 1.8 × 8.2 ±<br />
1.0 µm, n = 250/5], olivaceous or yellowish-brown, sometimes<br />
lighter coloured on one side; wall two-layered, ca. 0.5–1.3 µm<br />
thick, thinner on the lighter side, finely, densely verruculose,<br />
spore profile almost smooth or finely wavy.<br />
Additional specimens examined (paratypes): UK: Scotland: Outer<br />
Hebrides, Sgeir Ghlas Leac an Aiseig, Lewis, on Tractema verna,<br />
6 May 2010, P.A. Smith (KR 28182); Wales: Ceredigion, 100 m SW<br />
of mouth of Cwm Soden, on Tractema verna, 29 Apr. 2011, A.O.<br />
Chater (KRAM F-48878); Ceredigion, 200 m NE of Mwnt church,<br />
DISCUSSION<br />
The anther smuts of the genus Antherospora offer few<br />
morphological characteristics for inter-specific differentiation.<br />
This is the reason why they were formerly identified as a<br />
single species, Ustilago vaillantii (syn. Vankya vaillantii)<br />
(e.g. Vánky 1994). It appears that only a few species could<br />
be differentiated based on differences in spore sizes, and<br />
to a lesser extent also the spore wall thickness and the<br />
localization of the sori which is limited to the anthers or to the<br />
anthers and the surface of the inner floral organs (Bauer et<br />
al. 2008, Vánky et al. 2008, Vánky 2009). The recognition of<br />
Antherospora species that lack morphological differences is<br />
difficult or impossible without the support of molecular data.<br />
The variation of spore sizes between different collections<br />
of the same species (Bauer et al. 2008) may additionally<br />
complicate the situation with species delimitation. The<br />
differences in spore size range and mean spore size between<br />
different collections on Tractema verna (Table 1) confirm the<br />
variability of this character in Antherospora species, and it<br />
seems that whenever possible the morphology should be<br />
characterized based on collections from different populations.<br />
In spore size range (assessed from five specimens),<br />
the anther smut of Tractema verna is intermediate between<br />
Antherospora tourneuxii and A. urgineae on the one side<br />
and A. eucomis, A. scillae, A. vaillantii, and A. vindobonensis<br />
on the other side. Molecular data are not available for<br />
Antherospora eucomis, A. tourneuxii and A. urgineae.<br />
Other than infecting different host plant genera (Eucomis,<br />
Bellevalia and Charybdis respectively), A. eucomis can be<br />
separated by having smaller spores and a thinner spore<br />
wall, while the two remaining species have somewhat larger<br />
spores and thinner spore walls (Bauer et al. 2008, Vánky<br />
2009, Vánky et al. 2010). The spores of Antherospora<br />
scillae, A. vindobonensis (on Scilla) and A. vaillantii (on<br />
volume 2 · no. 2<br />
139
Piątek, Lutz, Smith & Chater<br />
ARTICLE<br />
Fig. 2. Antherospora tractemae (KRAM F-48879 – holotype). A. Type locality on Llangranog Head, Wales, United Kingdom. B. Flower of<br />
Tractema verna with infected anthers. C–I. Spores seen by LM, median and superficial views. Note somewhat lighter coloured and thinner one<br />
side of spores indicated by arrows on pictures F and H, and two-layered spore wall visible on picture E. J–M. Ornamentation of spores seen by<br />
SEM. Bars: B = 1 mm; C–J = 10 µm; K–M = 5 µm.<br />
Muscari) are smaller than those of the anther smut of<br />
Tractema verna, and the spore wall is additionally thinner<br />
in A. vaillantii. The spore wall thickness of Antherospora<br />
scillae and A. vindobonensis (0.8–1.5 µm, according to the<br />
key to Antherospora species by Vánky 2009) is comparable<br />
to those of the anther smut of Tractema verna, and different<br />
from all remaining Antherospora species that have spore<br />
walls of 0.5–0.8 µm thick. The molecular phylogenetic<br />
analyses separate the specimens on Tractema verna from<br />
these three Antherospora spp. (Fig. 1) and both ITS and<br />
LSU sequences differ significantly (Table 2). In conclusion,<br />
the morphology, the genetic difference, the results of<br />
the molecular phylogenetic analyses and different host<br />
plant genera support the recognition of the anther smut<br />
of Tractema verna as a new species, for which the name<br />
Antherospora tractemae is proposed in this study.<br />
140 ima fUNGUS
Antherospora tractemae sp. nov.<br />
The host plant was at first assigned to the genus Scilla (S.<br />
verna) according to most taxonomic treatments (e.g. McNeill<br />
1980), and thus it was initially assumed that its anther smut<br />
could be related to the two Antherospora species on Scilla<br />
already described. The molecular phylogenetic analyses<br />
revealed that the smut sporulating in anthers of Tractema<br />
verna occupies a position basal to the lineage of Muscariparasitizing<br />
Antherospora. A subsequent survey of the<br />
botanical literature revealed that Scilla is a polyphyletic genus<br />
and that Scilla verna actually belongs to the distinct genus<br />
Tractema (Speta 1998). Tractema verna has not as yet been<br />
included in phylogenetic analyses, but the related species<br />
Tractema monophyllos, the type of the genus Tractema,<br />
clusters distantly from the lineage attributed to Scilla s. str.<br />
(Pfosser & Speta 1999). Thus, the phylogenetic position of<br />
Antherospora tractemae supports the disentanglement of<br />
Scilla verna from Scilla s. str. This emphasizes the importance<br />
of the assignment of host species to the correct genus in<br />
order to promote the understanding of the evolutionary<br />
relationships between smut fungi and their host plants.<br />
Furthermore, it supports the long known hypothesis that<br />
plant parasitic fungi, especially rusts and smuts, can indicate<br />
relationships between their host plants (e.g. Savile 1954,<br />
1979, Nannfeldt 1968, Hijwegen 1979, 1988, Kukkonen &<br />
Timonen 1979).<br />
The large number of host plants reported for Ustilago<br />
vaillantii (syn. Vankya vaillantii) (Zundel 1953, Vánky<br />
1994) suggested that further species of Antherospora<br />
are likely to emerge. The descriptions of new species that<br />
lack morphological characteristics or have very subtle<br />
morphological differences needs to be supported by molecular<br />
data. The introduction of new species names based solely on<br />
supposed host specificity may be risky, because it is probable<br />
that within Antherospora there are species that have more<br />
than one host species as was evidenced for Antherospora<br />
vaillantii s. str. which is able to infect both Muscari comosum<br />
and M. neglectum (Bauer et al. 2008).<br />
ACKNOWLEDGEMENTS<br />
We thank Krzysztof Pawłowski (Kraków, Poland) and Michael Weiß<br />
(Tübingen, Germany) for translating the Latin description, Michael<br />
Weiß, Sigisfredo Garnica and Robert Bauer (Tübingen, Germany)<br />
for providing the molecular lab, Anna Łatkiewicz (Kraków, Poland)<br />
for help with the SEM pictures, and Roger G. Shivas (Indooroopilly,<br />
Australia) for helpful comments on the manuscript.<br />
REFERENCES<br />
Bauer R, Lutz M, Begerow D, Piątek M, Vánky K, Bacigálová<br />
K, Oberwinkler F (2008) Anther smut fungi on monocots.<br />
Mycological Research 112: 1297–1306.<br />
Bauer R, Lutz M, Piątek M, Vánky K, Oberwinkler F (2007)<br />
Flamingomyces and Parvulago, new genera of marine smut fungi<br />
(Ustilaginomycotina). Mycological Research 111: 1199–1206.<br />
Begerow D, Bauer R, Oberwinkler F (1997) Phylogenetic studies on<br />
nuclear LSU rDNA sequences of smut fungi and related taxa.<br />
Canadian Journal of Botany 75: 2045–2056.<br />
Begerow D, Stoll M, Bauer R (2006) A phylogenetic hypothesis<br />
of Ustilaginomycotina based on multiple gene analyses and<br />
morphological data. Mycologia 98: 906–916.<br />
Castresana J (2000) Selection of conserved blocks from multiple<br />
alignments for their use in phylogenetic analysis. Molecular<br />
Biology and Evolution 17: 540–552.<br />
Crous PW, Gams W, Stalpers JA, Robert V, Stegehuis G (2004)<br />
MycoBank: an online initiative to launch mycology into the 21 st<br />
century. Studies in Mycology 50: 19–22.<br />
Denchev CM, Giraud T, Hood ME (2009) Three new species of<br />
anthericolous smut fungi on Caryophyllaceae. Mycologia<br />
Balcanica 6: 79–84.<br />
Felsenstein J (1981) Evolutionary trees from DNA sequences: a<br />
maximum likelihood approach. Journal of Molecular Evolution<br />
17: 368–376.<br />
Gardes M, Bruns TD (1993) ITS primers with enhanced specificity for<br />
basidiomycetes – application to the identification of mycorrhizae<br />
and rusts. Molecular Ecology 2: 113–118.<br />
Gatesy J, DeSalle R, Wheeler W (1993) Alignment-ambiguous<br />
nucleotide sites and the exclusion of systematic data. Molecular<br />
Phylogenetics and Evolution 2: 152–157.<br />
Giribet G, Wheeler WC (1999) On gaps. Molecular Phylogenetics<br />
and Evolution 13: 132–143.<br />
Hijwegen T (1979) Fungi as plant taxonomists. Symbolae Botanicae<br />
Upsalienses 22(4): 146–165.<br />
Hijwegen T (1988) Coevolution of flowering plants with pathogenic<br />
fungi. In: Coevolution of Fungi with Plants and Animals (KA<br />
Pirozynski, DL Hawksworth, eds): 63–77. London: Academic<br />
Press.<br />
Huelsenbeck JP, Larget B, Miller RE, Ronquist F (2002) Potential<br />
applications and pitfalls of Bayesian inference of phylogeny.<br />
Systematic Biology 51: 673–688.<br />
Huelsenbeck JP, Rannala B (2004) Frequentist properties of<br />
Bayesian posterior probabilities of phylogenetic trees under<br />
simple and complex substitution models. Systematic Biology 53:<br />
904–913.<br />
Huelsenbeck JP, Ronquist F (2001) MRBAYES: bayesian inference of<br />
phylogenetic trees. Bioinformatics Applications Note 17: 754–755.<br />
Katoh K, Kuma K, Toh H, Miyata T (2005) MAFFT version 5:<br />
improvement in accuracy of multiple sequence alignment.<br />
Nucleic Acids Research 33: 511–518.<br />
Katoh K, Misawa K, Kuma K, Miyata T (2002) MAFFT: a novel<br />
method for rapid multiple sequence alignment based on fast<br />
Fourier transform. Nucleic Acids Research 30: 3059–3066.<br />
Katoh K, Toh H (2008) Recent developments in the MAFFT multiple<br />
sequence alignment program (outlines version 6). Briefings in<br />
Bioinformatics 9: 286–298.<br />
Kemler M, Lutz M, Göker M, Oberwinkler F, Begerow D (2009)<br />
Hidden diversity in the non-caryophyllaceous plant-parasitic<br />
members of Microbotryum (Pucciniomycotina: Microbotryales).<br />
Systematics and Biodiversity 7: 297–306.<br />
Kukkonen I, Timonen T (1979) Species of Ustilaginales, especially of<br />
the genus Anthracoidea, as tools in plant taxonomy. Symbolae<br />
Botanicae Upsalienses 22(4): 166–176.<br />
ARTICLE<br />
volume 2 · no. 2<br />
141
Piątek, Lutz, Smith & Chater<br />
ARTICLE<br />
Le Gac M, Hood ME, Fournier E, Giraud T (2007) Phylogenetic<br />
evidence of host-specific cryptic species in the anther smut<br />
fungus. Evolution 61: 15–26.<br />
Legon NW, Henrici A, Roberts PJ, Spooner BM, Watling R (2005)<br />
Checklist of the British and Irish Basidiomycota. Royal Botanic<br />
Gardens, Kew, Great Britain.<br />
Lutz M, Bauer R, Begerow D, Oberwinkler F, Triebel D (2004)<br />
Tuberculina, rust relatives attack rusts. Mycologia 96: 614–626.<br />
Lutz M, Göker M, Piątek M, Kemler M, Begerow D, Oberwinkler F<br />
(2005) Anther smuts of Caryophyllaceae: molecular characters<br />
indicate host-dependent species delimitation. Mycological<br />
Progress 4: 225–238.<br />
Lutz M, Piątek M, Kemler M, Chlebicki A, Oberwinkler F (2008)<br />
Anther smuts of Caryophyllaceae: molecular analyses reveal<br />
further new species. Mycological Research 112: 1280–1296.<br />
Lutz M, Vánky K, Bauer R (In press) Melanoxa, a new genus in the<br />
Urocystidales (Ustilaginomycotina). Mycological Progress. DOI<br />
10.1007/s11557-010-0737-7<br />
McNeill J (1980) Scilla L. In: Flora Europaea (TG Tutin, VH Heywood,<br />
NA Burges, DH Valentine, SM Walters & DA Webb, eds) 5: 41–<br />
43. Cambridge University Press.<br />
Nannfeldt JA (1968) Fungi as plant taxonomists. Acta Universitatis<br />
Upsaliensis 17: 85–95.<br />
O’Donnell KL (1992) Ribosomal DNA internal transcribed spacers<br />
are highly divergent in the phytopathogenic ascomycete<br />
Fusarium sambucinum (Gibberella pulicaris). Current Genetics<br />
22: 213–220.<br />
O´Donnell KL (1993) Fusarium and its near relatives. In: The fungal<br />
holomorph: mitotic, meiotic and pleomorphic speciation in fungal<br />
systematics. (DR Reynolds & JW Taylor, eds): 225–233. CAB<br />
International, Wallingford, United Kingdom.<br />
Pfosser M, Speta F (1999) Phylogenetics of Hyacinthaceae based<br />
on plastid DNA sequences. Annals of the Missouri Botanical<br />
Garden 86: 852–875.<br />
Posada D, Crandall KA (1998) MODELTEST: testing the model of<br />
DNA substitution. Bioinformatics 14: 817–818.<br />
Refrégier G, Le Gac M, Jabbour F, Widmer A, Shykoff JA, Yockteng<br />
R, Hood ME, Giraud T (2008) Cophylogeny of the anther smut<br />
fungi and their caryophyllaceous hosts: prevalence of host<br />
shifts and importance of delimiting parasite species for inferring<br />
cospeciation. BMC Evolutionary Biology 8: 100.<br />
Rodwell JS, Pigott CD, Ratcliffe DA, Malloch AJC, Birks HJB, et al.<br />
(1991–2000) British Plant Communities Vols 1–5. Cambridge<br />
University Press, Cambridge, UK.<br />
Ronquist FR, Huelsenbeck JP (2003) MRBAYES 3: Bayesian<br />
phylogenetic inference under mixed models. Bioinformatics 19:<br />
1572–1574.<br />
Savile DBO (1954) The fungi as aids in the taxonomy of the flowering<br />
plants. Science 120: 583–585.<br />
Savile DBO (1979) Fungi as aids to plant taxonomy: methodology<br />
and principles. Symbolae Botanicae Upsalienses 22(4): 135–<br />
145.<br />
Silvestro D, Michalak I (2010) raxmlGUI: a graphical front-end<br />
for RAxML. Available at http://sourceforge.net/projects/<br />
raxmlgui/.<br />
Speta F (1998) Systematische Analyse der Gattung Scilla L. s.l.<br />
(Hyacinthaceae). Phyton 38: 1–141.<br />
Stamatakis A (2006) RAxML-VI-HPC: maximum likelihood-based<br />
phylogenetic analyses with thousands of taxa and mixed models.<br />
Bioinformatics 22: 2688–2690.<br />
Stamatakis A, Hoover P, Rougemont J (2008) A rapid bootstrap<br />
algorithm for the RAxML web servers. Systematic Biology 57:<br />
758–771.<br />
Swofford DL (2001) PAUP* Phylogenetic Analysis Using Parsimony<br />
(*and Other Methods). Version 4. Sinauer Associates,<br />
Sunderland, Massachusetts, U.S.A.<br />
Vánky K (1994) European Smut Fungi. Stuttgart: G. Fischer Verlag.<br />
Vánky K (2009) Taxonomic studies on Ustilaginomycetes – 29.<br />
Mycotaxon 110: 289–324.<br />
Vánky K, Abbasi M, Samadi S (2010) Additions to the knowledge of<br />
smut fungi (Ustilaginomycetes) of Iran. Rostaniha 11(2): 191–<br />
198.<br />
Vánky K, Lutz M, Bauer R (2008) Floromyces, a new genus of<br />
Ustilaginomycotina. Mycotaxon 104: 171–184.<br />
White TJ, Bruns T, Lee S, Taylor J (1990) Amplification and direct<br />
sequencing of fungal ribosomal RNA genes for phylogenetics. In:<br />
PCR protocols, a guide to methods and applications. (MA Innis,<br />
DH Gelfand, JJ Sninsky & TJ White, eds): 315–322. Academic<br />
Press, San Diego, U.S.A.<br />
Zundel GL (1953) The Ustilaginales of the world. Pennsylvania State<br />
College School of Agriculture Department of Botany Contribution<br />
176: xi + 1–410.<br />
142 <br />
ima fUNGUS
doi:10.5598/imafungus.2011.02.02.05 <strong>IMA</strong> <strong>Fungus</strong> · volume 2 · no 2: 143–153<br />
Ascospore discharge, germination and culture of fungal partners of tropical<br />
lichens, including the use of a novel culture technique<br />
Ek Sangvichien 1 , David L. Hawksworth 2 , and Anthony J.S. Whalley 3<br />
ARTICLE<br />
1<br />
Faculty of Science, Ramkhamhaeng University, Hua Mark, Bangkok 10240, Thailand; corresponding author e-mail: bmsesang@yahoo.com<br />
2<br />
Departmento de Biología Vegetal II, Facultad de Farmacia, Universidad Complutense de Madrid, Plaza de Ramón y Cajal, Ciudad Universitaria,<br />
ES-28040 Madrid, Spain; and Department of Botany, Natural History Museum, Cromwell Road, London SW7 5BD, UK<br />
3<br />
School of Biomolecular Sciences, Liverpool John Moores University, Byrom Street, Liverpool L3 3AF, UK<br />
Abstract: A total of 292 lichen samples, representing over 200 species and at least 65 genera and 26 families,<br />
were collected, mainly in Thailand; 170 of the specimens discharged ascospores in the laboratory. Generally,<br />
crustose lichens exhibited the highest discharge rates and percentage germination. In contrast, foliose lichen<br />
samples, although having a high discharge rate, had a lower percentage germination than crustose species<br />
tested. A correlation with season was indicated for a number of species. Continued development of germinated<br />
ascospores into recognizable colonies in pure culture was followed for a selection of species. The most successful<br />
medium tried was 2 % Malt-Yeast extract agar (MYA), and under static conditions using a liquid culture medium, a<br />
sponge proved to be the best of several physical carriers tested; this novel method has considerable potential for<br />
experimental work with lichen mycobionts.<br />
Key words:<br />
Ascomycota<br />
colony development<br />
mycobiont<br />
seasonality<br />
Thailand<br />
Article info: Submitted 30 September 2011; Accepted 17 October 2011; Published 11 November 2011.<br />
Introduction<br />
The highest species diversity for most groups of organisms<br />
lies in the tropics. Lichenized fungi do not appear to be<br />
an exception, as Sipman & Aptroot (2001) estimated that<br />
between one-third and one-half of the world’s lichen diversity<br />
occurs there, and suggested that 50 % of the tropical<br />
lichen biota remained unknown. Yet there have been few<br />
experimental studies on ascospore discharge, germination,<br />
development of mycelia, and physiology of the fungal partners<br />
(mycobionts) of tropical lichens compared with those on<br />
temperate species. This is a major gap in our understanding<br />
of even basic aspects of the biology of tropical lichens.<br />
The first cited studies on the isolation of lichen-forming<br />
fungi are generally those of Töbler (1909) and Thomas<br />
(1939), although Töbler was primarily interested in the resynthesis<br />
of lichens from their individual symbionts (Turbin<br />
1996). However, Werner (1927), innovatively examined<br />
the effect of different media and additions on the growth of<br />
selected mycobionts from a range of lichens. Subsequent<br />
workers have concentrated on the development of methods<br />
for lichen re-synthesis (Ahmadjian 1964), and later Ahmadjian<br />
et al. (1980) and Ahmadjian & Jacobs (1981) produced the<br />
two most successful protocols (Bubrick 1988). Crittenden<br />
et al. (1995) were the first to attempt the isolation of a wide<br />
range of fungal partners of lichens, and also lichenicolous<br />
fungi, on a worldwide basis, although their material was<br />
predominantly from non-tropical regions. More recently,<br />
Yoshimura et al. (2002) reviewed the protocols available<br />
for isolation and cultivation of fungal and algal partners of<br />
lichens, emphasising studies by Japanese researchers, but<br />
again based largely on non-tropical material. A brief synopsis<br />
of methods used is provided by Stocker-Wörgötter & Hager<br />
(2008), with an emphasis on the production of extrolites<br />
(“lichen substances” or “secondary metabolites”).<br />
The lack of basic information on the isolation and<br />
growth of the fungal partners of tropical lichens provided the<br />
rationale for the present study. We investigated ascospore<br />
discharge from a wide range of tropical lichens in order to<br />
make a preliminary assessment of the conditions under<br />
which discharge occurred, and whether there could be any<br />
seasonal correlations. Observations on factors affecting<br />
germination and subsequent development on solid, or in<br />
liquid, growth media are also reported, since these are<br />
virtually undocumented for the fungal partners of tropical<br />
lichens. Our studies were carried out to identify apparent<br />
trends and <strong>issue</strong>s that merited in-depth investigations, as<br />
well as testing the efficacy of alternative culture methods.<br />
© 2011 International Mycological Association<br />
You are free to share - to copy, distribute and transmit the work, under the following conditions:<br />
Attribution:<br />
You must attribute the work in the manner specified by the author or licensor (but not in any way that suggests that they endorse you or your use of the work).<br />
Non-commercial: You may not use this work for commercial purposes.<br />
No derivative works: You may not alter, transform, or build upon this work.<br />
For any reuse or distribution, you must make clear to others the license terms of this work, which can be found at http://creativecommons.org/licenses/by-nc-nd/3.0/legalcode. Any of the above conditions can be waived if you get<br />
permission from the copyright holder. Nothing in this license impairs or restricts the author’s moral rights.<br />
volume 2 · no. 2 143
Sangvichien, Hawksworth & Whalley<br />
ARTICLE<br />
Table 1. Lichen collections according to locality and growth form type.<br />
Location Code Number of samples Crustose Foliose Erect shrubby or pendent<br />
Thailand<br />
Chiang Mai Province CM 27 15 11 1<br />
Chiang Rai Province CR 5 5 – –<br />
Kanjanaburi Province KJB 8 8 – –<br />
Nakon Sithammarat Province TS, RPB 6 6 – –<br />
Ratchaburi Province SP 7 7 – –<br />
Khao Yai National Park KY 208 140 60 8<br />
Phu Kradueng National Park PKD 5 4 – 1<br />
Sakaeraj Research Station SKR 2 2 – –<br />
Huai Kha Khaeng Wildlife Sanctuary HKK 4 4 – –<br />
Cambodia CAM 5 4 1 –<br />
Vietnam VN 15 15 – –<br />
Total 292 210 72 10<br />
Materials and Methods<br />
Taxon sampling<br />
The collection of samples began in 1998, and concentrated<br />
on Khao Yai National Park (KY), central Thailand. The<br />
remainder were collected during field surveys to Doi Suthep<br />
(18°49' N, 99°53' E) and Chiang Dao (l9°40' N, 99° E) in<br />
Chiang Mai Province (CM), Mae Fah Luang Arboretum (20°<br />
N, 99.5° E) Chiang Rai Province (CR), Sai Yok District (14°<br />
N, 99° E) Kanjanaburi Province (KJB), Khao Sok National<br />
Park (8° N, 99.5° E) Nakon Srithammarat Province (TS and<br />
RPB respectively), Phu Kradueng National Park, (16.8° N,<br />
101.8° E) Loei Province (PKD), Suan Phueng District (13.5°<br />
N, 99° E) Ratchaburi Province (SP), and Sakaeraj Research<br />
Station (14° N, 102° E) Nakhon Ratchasima Province (SKR).<br />
Some collections from Huai Kha Khaeng Wildlife Sanctuary<br />
Kanjanaburi Province, Vietnam (VN) and Cambodia (CAM)<br />
were donated by colleagues and friends. Khao Yai National<br />
Park was visited monthly during one year (1999–2000) for<br />
seasonal observations and experiments to explore the<br />
development of thalli, and also to ascertain if there were<br />
seasonal differences in ascospore discharge and spore<br />
viability<br />
Samples were cut into pieces, wrapped in t<strong>issue</strong> paper,<br />
and placed in individual strong brown paper bags. These<br />
were then returned to a survey house workroom and cleaned<br />
of attached soil or other extraneous material. Each sample<br />
was given a collection number, and information on the locality,<br />
substratum, and collection details were recorded. Then. if<br />
the specimens could not be immediately transferred to the<br />
laboratory for pre-isolation treatment, they were either kept<br />
in a cool place, or (where available) a domestic refrigerator,<br />
until they could be transferred. In the laboratory, samples<br />
were air-dried at room temperature (30 °C) overnight, and<br />
then transferred to new paper envelopes with identification<br />
labels and stored in a domestic refrigerator at 4 °C until the<br />
isolation protocol had been completed.<br />
Specimens were identified as precisely as possible on the<br />
basis of their morphology, anatomy, and chemical constituents<br />
(determined by standard thin-layer chromatographic methods;<br />
Orange et al. 2001). In many cases it was not possible to fully<br />
determine the samples to species as identification remains a<br />
major problem in tropical lichenology. The basic monographic<br />
treatments required to provide a sound taxonomic basis<br />
for studies of lichen distribution, ecology, and physiology<br />
are still lacking for most lichen families and genera in the<br />
tropics. Species that have not previously been described<br />
are also likely to be found; Homchantara & Coppins (2002)<br />
described 26 species of Thelotremataceae as new to science<br />
from Thailand 1 , and Aptroot et al. (2007) added 300 tropical<br />
species to the national list, of which 12 were new to science.<br />
The number of collections made for each morphological<br />
type of lichen, together with their geographical locations, are<br />
summarized in Table 1, while full details of selected collections<br />
for which positive results were obtained are given in Table 2.<br />
A list of the material collected is included as Supplementary<br />
Information (Table S1, online only) and in Sangvichien (2005).<br />
Voucher specimens are maintained in The Lichen Herbarium,<br />
Ramkhamhaeng University, Bangkok (RAMK).<br />
Spore discharge and germination<br />
The specimens were removed from storage, and surfacecleaned<br />
with air from an aerosol camera blower to remove<br />
any remaining soil and debris. A sterile surgical blade<br />
(Gowlands No. 11) was used to dissect specimens to obtain<br />
small portions with ascomata, and the remainder of the<br />
samples were then returned to storage. The process was<br />
repeated if the first isolation attempts were unsuccessful. The<br />
portions of lichen with ascomata, or occasionally only a single<br />
ascoma, were attached with a small quantity of petroleum<br />
jelly onto the inverted lid of a 9 cm diam Petri dish (Sterilin).<br />
The spores were allowed to shoot upwards onto an overlying<br />
layer of Tap Water Agar (WA; Booth 1971). The agar surface<br />
was examined daily with a stereozoom binocular microscope<br />
(Olympus SZ11), and once ascospores had been discharged,<br />
1<br />
Eleven of these have since proved to be synonyms of previously<br />
described species (Papong et al. 2010).<br />
144 ima fUNGUS
Tropical lichen fungi: ascospore discharge and culture<br />
small agar blocks (3–5 mm 2 ) with ascospores on the surface<br />
were excised and transferred to Malt-Yeast extract Agar<br />
(MYA; Merck or Oxoid). Germination of ascospores was<br />
assessed under the stereozoom microscope; observations<br />
were made daily for 7 d, and subsequently twice weekly.<br />
If no germ tubes had been observed after six weeks, then<br />
“no-germination” in that collection was recorded. Germinated<br />
ascospores were maintained at room temperature for further<br />
studies on growth and colony morphology, or were used as<br />
inoculum for liquid media.<br />
In order to investigate the seasonality and discharge of<br />
ascospores, thalli were collected each month from the same<br />
trees in Khao Yai National Park over a one year period,<br />
and their discharge patterns and rates of germination were<br />
determined for each monthly sample following the protocol<br />
described above.<br />
The distance to which ascospores were discharged was<br />
studied in a representative sample of 15 species. Clear<br />
plastic boxes 18 x 7.5 x 5 cm were used with a layer of tap<br />
water agar in the lid of the box. Ascomata samples, approx.<br />
0.3 mm diam, were attached to one vertical microscope<br />
slide (2 x 4 cm) which was shallowly immersed in the agar<br />
layer. The box was then incubated on the laboratory bench<br />
at an ambient temperature of 25–30 °C, with approximately<br />
12 h of daylight. The agar surface was examined under an<br />
Olympus stereozoom microscope (Model SZ 11) daily for 5 d.<br />
If no spores were discharged within 3 d, the procedure was<br />
repeated, and then, if after a second 3 d period no discharge<br />
was observed, the protocol was repeated for a third and final<br />
time.<br />
We also investigated the effects of relative humidity by<br />
incubating Petri dishes in plastic moist chambers containing<br />
different saturated solutions to maintain the relative humidity<br />
at particular levels, following Kaye & Laby (1966): potassium<br />
nitrate (92 %), ammonium sulphate (80 %), and sodium<br />
nitrate (65 %).<br />
We tried, but did not adopt, the surface sterilization<br />
protocol of Crittenden et al. (1995) as we found it to be<br />
detrimental to ascospore discharge in the tropical lichens<br />
tested; in consequence, untreated lichen samples were used<br />
throughout.<br />
pieces of the fungal cultures into the liquid medium, different<br />
types of physical support for the fungi on the surface of the<br />
liquid were tested.<br />
Four types of material were evaluated: (1) Stacked<br />
Membrane filters (pores 0.22 μm diam; polyvinylidene<br />
fluoride, PVDF) were promising when tested first, but the<br />
slippery surface when floating on the liquid rendered them<br />
difficult to inoculate. (2) Whatman No.1 filter papers were<br />
tested in order to overcome the problem of stacked layers. (3)<br />
Kraft paper was tried as an alternative to Whatman No.1. And<br />
(4), synthetic sponge (polystyrene) pieces 2.5 x 2.5 x 0.3 cm,<br />
together with pieces of fungal colonies cut from solid media<br />
of 0.4 x 0.4 cm, placed on the surface of these materials, and<br />
floated on the surface of 50 ml MYB in 250 mL Erlenmeyer<br />
flasks. Observations were made twice daily and, at the same<br />
time, the flasks were gently swirled for 10–15 s to circulate<br />
the medium.<br />
Since poor aeration could be a factor limiting growth, the<br />
effect of increased aeration was tested in two ways. First, air<br />
was supplied by an aquarium air pump and passed through<br />
a sterile filter (Sartorious, Sartofluor ® ; pores 0.2 µm diam) to<br />
prevent contamination. Second, flasks were placed on an<br />
orbital shaker (Innova 4230, New Brunswick). Shake cultures<br />
were prepared using inocula produced in the same way as for<br />
static cultures, and transferred to 250 mL Erlenmeyer flasks<br />
containing 50 mL of MYB. The orbital shaking incubator was<br />
set at a speed of 200 rpm, and at a temperature of 30 ± 0.5<br />
°C.<br />
Scanning electron microscopy<br />
Specimens for scanning electron microscopy (SEM), either<br />
intact ascomata or cultures of the isolated fungal partners,<br />
were fixed in 5 % glutaraldehyde and dehydrated in a graded<br />
ethyl alcohol series. The specimens were then attached to<br />
aluminium stubs using either Dag metallic paint or adhesive<br />
carbon pads to prevent electron charging of the specimens.<br />
The samples were gold-coated using a Sputter Coating Unit<br />
(Polaron RU-SC7620) and examined either with a Jeol 840<br />
SEM, a Jeol SEM5410LV, or a Leo 1455VP scanning electron<br />
microscope. Digital images were produced using an imagecapture<br />
system (Röentec) or with accessories of Leo 1455 VP.<br />
ARTICLE<br />
Fungal culture on solid media<br />
MYA (see above) was the medium of choice for all cultures<br />
of the fungal partners, but Potato Dextrose Agar (PDA),<br />
Corn Meal Agar (CMA), Oatmeal Agar (OMA), and Czapek-<br />
Dox Agar (CDA), were also used to determine the optimum<br />
medium for growth. For recipes see Booth (1971).<br />
Fungal culture in liquid media<br />
Malt Yeast Extract Broth (MYB) was selected as the standard<br />
medium for studies of the isolated fungi in liquid culture,<br />
since good growth rates of several fungal partners had been<br />
observed on solid MYA. MYB has also been favoured by<br />
previous researchers (e.g. Hamada 1989, Honegger 1990,<br />
Yamamoto et al. 1998). Static culture was most frequently<br />
used, and following initial trials with direct inoculation of<br />
Results and Discussion<br />
This study employed a large number of samples in order to<br />
gain an impression of possible general features of ascospore<br />
discharge and development in tropical lichens to provide a<br />
basis on which to determine directions future investigations<br />
might take. As replicates were not used for most of the<br />
species, the conclusions must be viewed as preliminary<br />
and treated with caution. Nevertheless, some indications of<br />
trends emerged, although we recognize that further work may<br />
require their modification or refinement. This caveat must be<br />
borne in mind with respect to this discussion of our results.<br />
The number of crustose lichen collections made was<br />
much larger than that of foliose lichens (Table 1). Crustose<br />
volume 2 · no. 2<br />
145
Sangvichien, Hawksworth & Whalley<br />
ARTICLE<br />
Table 2. Lichen collections, ascospores discharged, germination (%), and colony development in selected species. The classification follows<br />
Lumbsch & Huhndorf (2010).<br />
Taxon Collection number Ascospores discharged Germination (%) Colony development<br />
ARTHONIALES<br />
Arthoniaceae<br />
Arthothelium sp. KY175 11 100 2<br />
LECANORALES<br />
Cladoniaceae<br />
Cladonia submultiformis KY117,118,119 > 500 100 1<br />
Haematommataceae<br />
Haematomma puniceum KY107 NA ND 0<br />
Haematomma sp. VN3 NA ND 0<br />
Lecanoraceae<br />
Lecanora cenisia SP4 47 0 0<br />
Lecanora intumescens CM27 NA ND +<br />
Lecanora leprosa SP6 310 0 0<br />
Lecanora polytropa KY177 47 92 0<br />
Pyrrhospora sp. 1 CR8 NA ND +<br />
Parmeliaceae<br />
Parmelina sp. CM33 50 0 0<br />
Relicinopsis sp. KY81 NA ND 0<br />
Relicinopsis sp. CM32 191 0 0<br />
Usnea complanata CM12 > 500 0 0<br />
Pilocarpaceae<br />
Sporopodium argillaceum VN16 2 50 +<br />
Ramalinaceae<br />
Bacidia subannexa SP10 > 900 89 0<br />
Bacidia sp. 1 SP12 190 21 0<br />
OSTROPALES<br />
Graphidaceae<br />
Cyclographina sp. 2 KY390 23 57 0<br />
Glyphis cicatricosa SP7 12 100 3<br />
Glyphis cicatricose KY231 NA ND +<br />
Graphina cleistoblephara KY129 96 98 +<br />
Graphina hiascens KY160 69 99 3<br />
Graphina sp. 2 KY104 NA ND +<br />
Graphina sp. 5 KY157 124 91 +<br />
Graphina sp. 9 KY124 10 90 +<br />
Graphina sp.18 KY171 28 100 3<br />
Graphina sp. 19 KY91 NA ND +<br />
Graphina sp. 20 KY180 186 90 +<br />
Graphis afzelii CR5 NA ND +<br />
Graphis albocolpata KY147 NA ND +<br />
Graphis analoga HKK7 > 1000 100 0<br />
Graphis apertella HKK4 255 100 0<br />
Graphis apertella SP2 307 259 0<br />
Graphis elegans KY162 73 99 3<br />
Graphis kakaduensis TS3 435 100 0<br />
Graphis librata RPB1 760 100 0<br />
146 ima fUNGUS
Tropical lichen fungi: ascospore discharge and culture<br />
Table 2. (Continued).<br />
Taxon Collection number Ascospores discharged Germination (%) Colony development<br />
Graphis rigidula KY165 > 500 100 3<br />
Graphis rimulosa CR3 5 100 3<br />
Graphis xanthospora TS2 56 100 0<br />
Graphis sp. 10 KY148 111 94 3<br />
Graphis sp. KY133 5 0 0<br />
Graphis rimulosa SP1 180 100 0<br />
Graphis rimulosa KY133 NA ND +<br />
Graphis sp. SP9 > 1000 100 3<br />
Gyrostromum sp. KY161 26 96 +<br />
Ocellularia s. lat. sp. KY173 129 89 3<br />
Phaeographina caesioradians HKK1 450 100 0<br />
Phaeographina quassiaecola VN6 1 100 3<br />
Phaeographina sp. CAM5 1 100 3<br />
Phaeographina sp. 4 HKK2 > 1000 100 3<br />
Pheopgraphis melanostalazans KY144 113 88 3<br />
Phaeographis melanostalazans KY121 NA ND +<br />
Phaeographis pyrhochora PKD4 42 100 3<br />
Phaeographis sp. 27 KY229 NA ND +<br />
Sarcographa actinobola KY205 NA ND +<br />
Sarcographa labyrinthica KY240 NA ND +<br />
Thelotrema s. lat. sp. 3 KY233 NA ND +<br />
Thelotrema sp. 4 VN9 1 0 0<br />
Thelotrema s. lat. sp. 5 KY245 NA ND +<br />
Thelotrema sp. 6 VN10 1 100 +<br />
ARTICLE<br />
PELTIGERALES<br />
Nephromataceae<br />
Nephroma sp. CM30 15 67 0<br />
PERTUSARIALES<br />
Pertusariaceae<br />
Pertusaria sp. 4 PKD2 18 100 2<br />
PYRENULALES<br />
Pyrenulaceae<br />
Pyrenula sp. KY208 NA ND +<br />
Pyrenula sp. KY230 NA ND +<br />
Pyrenula sp. KY249 NA ND +<br />
Pyrenula sp. KY95 NA ND +<br />
TELOSCHISTALES<br />
Physciaceae<br />
Rinodina sp. KY169 NA ND +<br />
Teloschistaceae<br />
Caloplaca sp. CR6 2 0 0<br />
TRYPETHELIALES<br />
Trypetheliaceae<br />
Campylothelium sp. SKR2 NA NA +<br />
volume 2 · no. 2<br />
147
Sangvichien, Hawksworth & Whalley<br />
ARTICLE<br />
Table 2. (Continued).<br />
Taxon Collection number Ascospores discharged Germination (%) Colony development<br />
Laurera benguelensis KY61 197 99 3<br />
Laurera madreporiformis KY14 NA ND +<br />
Laurera megasperma KY238 NA NA 3<br />
Laurera meristospora KY195 NA NA 3<br />
Laurera subdiscreta SKR1 NA NA +<br />
Pseudopyrenula diluta KY113 NA ND +<br />
Trypetheliaceae sp. KY135 39 100 3<br />
Trypetheliaceae sp. KY131 63 87 +<br />
Trypethelium eluteriae KY66 82 100 3<br />
Trypethelium ochroleucum KY235 NA ND +<br />
INCERTAE SEDIS<br />
Unidentified KY141 46 83 +<br />
Unidentified KY143 6 50 +<br />
NA = not available, ND = not detected.<br />
Colony development : 0 = none, 1 = poor, 2 = moderate, 3 = good , + = germination but with premature cessation of growth.<br />
Fig. 1. Ascospore germination in selected species. A. Glyphis<br />
cicatricosa (KY231). B. Phaeographina montagnei (KY263). C–D.<br />
Pyrenula sp. (KY95 and 208). E–F. Trypethelium eluteriae (KY66).<br />
G-H. Trypethelium tropicum (KY131).<br />
lichens were preferentially selected for experimentation<br />
as preliminary studies suggested that their ascospores<br />
germinated more readily on artificial media. Ascomata of<br />
erect shrubby (fruticose) and pendent lichens were much less<br />
common than crustose or foliose ones, especially at Khao Yai<br />
National Park, and so could not be investigated further.<br />
Of the 292 lichen samples collected (Table 1), 170<br />
samples liberated ascospores in the laboratory, and in several<br />
instances successive samples of the same lichen exhibited<br />
high percentage germination rates (Table 2, Fig.1). Crustose<br />
lichens exhibited the highest rate of spore discharge, and also<br />
subsequent germination. In contrast, foliose lichen species<br />
(e.g. Heterodermia diademata) exhibited a high discharge<br />
rate, but only a low percentage of ascospores germinated.<br />
Seasonal influences on ascospore discharge and<br />
germination were explored in selected species (Fig. 2). In<br />
Trypethelium eluteriae (KY 66), Graphis elegans (KY162),and<br />
G. rigidula (KY165), ascospores were discharged readily<br />
each month, and spores from each monthly sample also<br />
germinated. However, in contrast, Cladonia submultiformis<br />
(KY117) discharged ascospores only towards the end of the<br />
winter (January-February), and in the summer (April-June)<br />
none were discharged (Fig. 2).<br />
Following germination, the ability of the isolated fungi to<br />
continue to develop and form colonies was investigated. In<br />
some common crustose lichen species, the fungal partners<br />
grew well and produced small colonies within a few months<br />
(Fig. 3). Species of Trypethelium and Laurera developed<br />
colonies readily, while in Haematomma wattii and Lecanora<br />
intumescens the spores germinated but growth was either<br />
very slow or soon ceased. In addition to growth on solid<br />
media, liquid culture under static growth conditions was<br />
tried, but generally growth was slow. Further, when the<br />
fungi were inoculated onto the surface of membrane filters<br />
floating on the surface of MYB, this was not satisfactory as<br />
148 ima fUNGUS
Janu<br />
Febua<br />
Mar<br />
A<br />
M<br />
Ju<br />
J<br />
Augu<br />
Septemb<br />
Octob<br />
Novem<br />
Decemb<br />
%<br />
Janua<br />
January<br />
Febua<br />
Febuary Febua<br />
March<br />
Mar<br />
Mar<br />
Ap<br />
April A<br />
M<br />
May<br />
Ju<br />
June Ju<br />
July<br />
J<br />
Tropical lichen fungi: ascospore discharge and culture<br />
Months<br />
0<br />
Months<br />
August<br />
Augu<br />
Augu<br />
September<br />
Septemb<br />
Octob<br />
October Octob<br />
November<br />
Novem<br />
December<br />
Decemb<br />
0<br />
January Janua<br />
Febuary Febua<br />
March Mar<br />
A<br />
April<br />
Months<br />
% spore % spore germination<br />
100<br />
100 80<br />
80 60<br />
60 40<br />
40 20<br />
200<br />
0<br />
January January<br />
Febuary Febuary<br />
March March<br />
April<br />
April<br />
B<br />
A<br />
May<br />
June<br />
July<br />
May<br />
June<br />
July<br />
Months<br />
August August<br />
September September<br />
October October<br />
November November<br />
December December<br />
% spore spore % spore<br />
%<br />
germination<br />
spore germination<br />
100<br />
100 80<br />
100<br />
80<br />
100 60 80<br />
60 60<br />
40 80<br />
60 40<br />
40<br />
20<br />
40 20<br />
20<br />
20 00<br />
00<br />
January<br />
January<br />
January<br />
January<br />
Febuary<br />
Febuary<br />
Febuary<br />
Febuary<br />
March<br />
March March<br />
March<br />
April<br />
April<br />
April<br />
April<br />
May<br />
May<br />
May<br />
May<br />
C<br />
B<br />
June<br />
June<br />
June<br />
June<br />
July<br />
July<br />
July<br />
July<br />
Months<br />
August<br />
August August<br />
August<br />
September<br />
September September<br />
September<br />
October October<br />
October October<br />
November<br />
November<br />
November<br />
November<br />
December<br />
December<br />
December<br />
December<br />
% spore % spore germination<br />
100<br />
100<br />
80<br />
80<br />
60<br />
60<br />
40<br />
40<br />
20<br />
20<br />
0<br />
0<br />
January January<br />
Febuary Febuary<br />
ARTICLE<br />
March March<br />
April April<br />
Months<br />
Months<br />
% spore % spore germination<br />
100<br />
100 80<br />
80 60<br />
60 40<br />
40 20<br />
200<br />
0<br />
January January<br />
Febuary Febuary<br />
March March<br />
April<br />
Fig. 2. Apparent seasonal effect on ascospore D discharge and germination on 100 selected species. A. Cladonia submultiformis (KY117). B. Graphis<br />
elegans (KY162). C. G. rigidula (KY165). C D. Trypethelium eluteriae (KY66). 100 80<br />
D<br />
100<br />
% spore % spore germination<br />
100 80<br />
80 60<br />
the membranes 60 40<br />
often sank following inoculation, or tended to<br />
40 20<br />
collapse after a period of growth. Amongst the other physical<br />
200<br />
carriers tested, were segments of unbleached Kraft paper or<br />
0<br />
Whatman filter paper several layers thick. Growth occurred<br />
on the surface of these, but it was only possible to assess<br />
Months<br />
this visually as it proved impossible to physically separate<br />
Months<br />
all the fungal material from their surfaces. In contrast, the<br />
sponge pieces tested as alternative carriers facilitated growth<br />
after incubation in the fungal partners D tested; in most cases,<br />
sponge proved to be superior to the other carriers tried<br />
(Fig. 3).<br />
Trypethelium 100 eluteriae (KY66).<br />
80<br />
No contamination by spores from other fungi during<br />
60<br />
discharge were 40 encountered; the distinctive ascospores<br />
from the lichens 20 were deposited, either as small packets of<br />
0<br />
spores or as single spores, and could easily be recognized<br />
for subculturing using a stereozoom microscope.<br />
Our results suggest that high spore discharge rates are<br />
Months<br />
correlated with the freshness of the samples and season of<br />
collection, as well as the state of maturity of the ascomata.<br />
Spore germination also appeared to be correlated with<br />
species distributions. Widely distributed species, such as<br />
Trypethelium eluteriae, Laurea bengualensis, and most<br />
Graphidaceae Trypethelium studied, eluteriae exhibited (KY66). relatively high rates of<br />
germination.<br />
There was, however, considerable variation in ascospore<br />
discharge between the species tested, and, also between<br />
different collections of the same species. In Laurera<br />
bengualensis and L. madreporiformis, spores were readily<br />
discharged in all of the collections examined, but in L.<br />
meristospora, although discharge occurred, it was at a much<br />
lower rate. In Trypethelium eluteriae, spore discharge occurred<br />
% spore germination<br />
100<br />
January January<br />
January<br />
Febuary Febuary<br />
Febuary<br />
March March<br />
March<br />
April<br />
April<br />
April<br />
April<br />
May<br />
May<br />
May<br />
May<br />
May<br />
C<br />
B<br />
June<br />
June<br />
July<br />
Months<br />
June<br />
June<br />
July<br />
Months<br />
June<br />
July<br />
July<br />
July<br />
August August<br />
August August<br />
August<br />
September September<br />
September September<br />
September<br />
October October<br />
October October<br />
October<br />
November November<br />
November November<br />
November<br />
December December<br />
December December<br />
December<br />
% spore %<br />
spore<br />
spore<br />
%<br />
germination<br />
spore germination<br />
% spore<br />
% spore %<br />
germination<br />
spore germination<br />
100 80<br />
100<br />
80<br />
100 60 80<br />
60 60<br />
40 80<br />
60 40<br />
40<br />
20<br />
40 20<br />
20<br />
20 00<br />
00<br />
60 40 80<br />
January<br />
January<br />
January<br />
January<br />
Febuary<br />
Febuary<br />
Febuary<br />
Febuary<br />
March<br />
March<br />
March<br />
March<br />
April<br />
April<br />
May<br />
May<br />
June<br />
June<br />
July<br />
July<br />
Months<br />
throughout 60<br />
40 the year, but in Cladonia submultiformis, although<br />
20<br />
40<br />
40<br />
ascospores were also readily discharged, only those from the<br />
20<br />
20 0<br />
20<br />
end of the winter season (February) germinated. This suggests<br />
00<br />
0<br />
that in some lichen-fungi, seasonality is important even in the<br />
tropics. These observations of differences in spore discharge<br />
Months<br />
between species are in agreement with those of Crittenden et<br />
Months<br />
al. (1995), based mainly on samples from non-tropical regions.<br />
The distance of discharge of ascospores is important for<br />
the dissemination of the species (Table 3). Of the 15 species<br />
100<br />
tested, those of Graphina sp. 9 (KY104) were discharged the<br />
80<br />
furthest, to 63 mm. This figure compares with the maximum of<br />
60<br />
45 mm obtained for the temperate Rhizocarpon umbilicatum<br />
40<br />
(Bailey & Garrett 1968). However, in that study no information<br />
20<br />
was given as to the distance attained by the majority of spores,<br />
0<br />
which is perhaps the most pertinent parameter in relation to<br />
effective dispersal and establishment in nature. There was a<br />
wide variation even within the 75 % ranges of projection in<br />
Months<br />
many cases, and we speculate that this could be an adaptation<br />
to increase the probability of contact with a suitable new<br />
substrate. In nature, air turbulence currents and wind would<br />
also be expected to influence the final distance travelled.<br />
Environmental conditions also appeared to influence<br />
ascospore discharge. At 15 °C any discharge was rare,<br />
occurring only in Arthopyrenia sp. (PKD3) and Laurera<br />
subdiscreta (SKR1), while at 45 °C no discharge was<br />
observed in any species tested (Table 4). As might have been<br />
expected for tropical species, most species discharged at 30<br />
°C and 35 °C, but the mean optimum discharge temperature<br />
for all species was close to 25 °C. Relative humidity also<br />
appears to be important, with three of the four species<br />
investigated discharging at relative humidities from 65 %<br />
August<br />
August<br />
August<br />
August<br />
80<br />
Trypethelium 100 60 eluteriae (KY66).<br />
Fig. 2. Apparent seasonal effect on ascospore discharge and germination on selected species. A.<br />
Cladonia submultiformis (KY117). B. Graphis elegans (KY162). C. G. rigidula (KY165). D.<br />
% spore germination<br />
100<br />
January<br />
January<br />
January<br />
January<br />
Febuary<br />
Febuary<br />
Febuary<br />
Febuary<br />
March<br />
March<br />
March<br />
March<br />
April<br />
April<br />
April<br />
April<br />
April<br />
May<br />
May<br />
May<br />
May<br />
May<br />
D<br />
C<br />
June<br />
June<br />
June<br />
June<br />
June<br />
July<br />
July<br />
Months<br />
July<br />
July<br />
July<br />
August<br />
August<br />
August<br />
August<br />
September<br />
September<br />
September<br />
September<br />
Fig. 2. Apparent seasonal effect on ascospore discharge and germi<br />
Cladonia submultiformis (KY117). B. Graphis elegans (KY162). C<br />
Fig. 2. Apparent seasonal effect on ascospore discharge and germination on selected species. A.<br />
Fig. 2. Apparent seasonal effect on ascospore discharge and germi<br />
Cladonia submultiformis (KY117). B. Graphis elegans (KY162). C. G. rigidula (KY165). D.<br />
Cladonia submultiformis (KY117). B. Graphis elegans (KY162). C<br />
Trypethelium eluteriae (KY66).<br />
April<br />
May<br />
June<br />
July<br />
September<br />
September<br />
September<br />
September<br />
October October<br />
October October<br />
October October<br />
October<br />
October<br />
November<br />
November<br />
November<br />
November<br />
November<br />
November<br />
November<br />
November<br />
December<br />
December<br />
December<br />
December<br />
December<br />
December<br />
December<br />
December<br />
% spore % spore germination<br />
% spore germination<br />
100<br />
100 80<br />
80<br />
60<br />
60 40<br />
40<br />
20<br />
200<br />
0<br />
80<br />
60<br />
January January<br />
January<br />
Febuary Febuary<br />
Febuary<br />
March March<br />
March<br />
April<br />
April<br />
April<br />
volume 2 · no. 2<br />
149
Sangvichien, Hawksworth & Whalley<br />
ARTICLE<br />
Fig. 3. Development and growth of selected species. A. SEM of germinating ascospore of Cyclographina platyleuca (KY390). B. SEM of<br />
germinating ascospore of Phaeographina chapadana (KY474). C. Colony development of Thelotrema sp. on 2 % MYA agar (VN10). D. Colony<br />
development of Sarcographa labyrinthica (KY240). E. 2 % MYB broth showing colony development of Trypethelium eluteriae (KY131) under<br />
static conditions. F–H. Colony of T. eluteriae (KY131) developing on sponge in 2 % MYB broth under static conditions. Bars: A = 10 µm; E = 3<br />
cm; F, H = 1 cm.<br />
150 ima fUNGUS
Tropical lichen fungi: ascospore discharge and culture<br />
Table 3. Ascospore discharge and distance of projection from selected lichens studied (arranged in descending order).<br />
Species<br />
Collection<br />
number<br />
Number of ascopores<br />
discharged<br />
Distance ascospores projected Minimum – (average<br />
range of 75 % spores) – Maximum (mm)<br />
Graphina sp. 2 KY104 52 5–(10–50)–63<br />
Thelotrema s. lat. sp. 3 KY233 114 7.5–(10–29)–40<br />
Graphina sp. 20 KY180 78 2.5–(3.5–20)–38<br />
Graphina hiascens KY160 38 14.5–(15–21.5)–36<br />
Graphina sp. 10 KY217 57 15–(15–31)–34.5<br />
Sarcographa actinobola KY205 62 2.5–(5–25)–26<br />
Graphis elegans KY162 154 3–(6–15)–24.5<br />
Glyphis cicatricosa KY231 183 3.5–(7.5–21)–24<br />
Pyrenula sp. 6 KY206 81 8–(10–15)–21.5<br />
Pheographis sp. 27 KY229 67 5–(9–15)–21.5<br />
Graphina sp. 9 KY124 4 12.5–20<br />
Trypethelium ochrolecum KY235 19 4–(4–10)–18<br />
Sarcographa labyrinthica KY240 102 3.5–(3–15)–18<br />
Graphis albocolpata KY147 95 5–(5–9.5)–14.5<br />
Buellia sp. 5 KY220 31 1–(1–9.5)–12.5<br />
ARTICLE<br />
Table 4. Ascospore discharge from selected species over the temperature range 15–45 °C.<br />
Species<br />
Collection 15 °C 20 °C 25 °C 30 °C 35 °C 45 °C<br />
number<br />
Arthopyrenia sp. PKD3 + + + + + –<br />
Graphina sp. 9 KY124 – + + + – –<br />
Graphis albocolpata KY147 – – + + + –<br />
Haematomma puniceum KY108 – + + – + –<br />
Laurera subdiscreta SKR1 + + + + – –<br />
Pertusaria sp. 4 PKD2 – + + – + –<br />
Phaeographina pyrrhochroa PKD4 – + + – + –<br />
Phaeographina sp. CAM5 – + + + – –<br />
Pyrrhospora sp. 1 PKD1 – + + + + –<br />
Trypethelium eluteriae KY79 – + + + – –<br />
to 100 % (Fig. 4). However, Laurera subdiscreta (SKR1),<br />
L. benguelensis (KY 61), Pyrenula sp. (KY95), and Graphis<br />
sp. 3 (KY260), discharged only at 100 % relative humidity.<br />
These results suggest that both temperature and relative<br />
humidity, which will vary with habitat and season, influence<br />
ascospore discharge in tropical lichens to different degrees,<br />
something that would be a major factor in their performance<br />
and occurrence in nature.<br />
Ascospore germination appeared also to be linked to<br />
species distributions. Widely distributed species such as<br />
Trypetheium eluteriae and Laurera bengualensis, together<br />
with most Graphidaceae tested, exhibited relatively high rates<br />
of ascospore germination. Ascospores of crustose lichens<br />
generally germinated readily, whilst those from shrubby and<br />
pendent species were much more difficult, or failed to germinate.<br />
Ascospores of the different fungi exhibited several<br />
distinctive germination patterns (Fig.1A–H): (1) multiple<br />
germination tubes developing from different regions of the<br />
spore (e.g. Pyrenula and Arthopyrenia species, Graphis<br />
cicatricosa, Laurera benguelensis, Graphina irabensis; (2)<br />
bipolar germination (e.g. Trypethelium tropicum); and (3)<br />
multiple germination tubes developing all over the spore from<br />
individual segments within them (e.g. Thelotremataceae,<br />
Cyclographina platyleuca KY390/RPB3).<br />
When germination was successful, fungal partners of<br />
most crustose species tested grew well on solid media, with<br />
small colonies developing within a few months. Trypethelium<br />
and Laurera species generally grew well, but Haematomma<br />
wattii and Lecanora intumescens developed very slowly<br />
and growth often ceased – even though the ascospores<br />
germinated readily. Growth in liquid culture was generally<br />
very slow, and static culture was found to be superior to<br />
shake culture for all species tested. However, growth on static<br />
liquid culture was much enhanced by the use of a physical<br />
carrier. While segments of Kraft paper or Whatman filter<br />
paper proved to be successful carriers, sponge pieces were<br />
superior in relation to visible enhanced growth. We consider<br />
that sponge pieces used as a carrier have a wide potential<br />
for studies on the physiology and development of lichen<br />
fungi as the colonies can be transferred without disruption<br />
volume 2 · no. 2<br />
151
Sangvichien, Hawksworth & Whalley<br />
ARTICLE<br />
Spore germination (%)<br />
100<br />
80<br />
60<br />
40<br />
20<br />
A<br />
Spore germination (%)<br />
100<br />
80<br />
60<br />
40<br />
20<br />
B<br />
0<br />
1 2 3 4 5<br />
Days<br />
0<br />
1 2 3 4 5<br />
Days<br />
C<br />
D<br />
100<br />
100<br />
Spore germination<br />
80<br />
60<br />
40<br />
20<br />
Spore germination (%)<br />
80<br />
60<br />
40<br />
20<br />
0<br />
1 2 3 4 5<br />
Days<br />
0<br />
1 2 3 4 5<br />
Days<br />
Fig. 4. Ascospore discharge and germination in selected species as influenced by percentage humidity in the experiments. A. Laurera subdiscreta<br />
(SKR1). B. L. benguelensis (KY61). C. Graphis sp. 3 (KY 260). D. Pyrenula sp. (KY95).<br />
to different liquid media. This means that, for example, the<br />
effect of different nutrients in the medium on the production<br />
of extrolites could be explored. Culberson & Armaleo<br />
(1992), in their investigation of Cladonia grayi, previously<br />
concluded that the production of compounds concentrated<br />
in the naturally occurring lichen was linked to the aerial<br />
growth habit. Their conclusion was based on the finding that,<br />
following the transfer of lightly fragmented mycelia from liquid<br />
to solid media, there was a subsequent proliferation of aerial<br />
hyphae and extrolite production. Although only a limited<br />
investigation of the chemical products of the isolated fungal<br />
partners of the tropical lichens was undertaken in our study,<br />
comparison of extrolites from the whole lichen thallus with<br />
those produced by the fungal partner alone indicated that in<br />
some cases more compounds were produced by the whole<br />
thallus than in the isolated fungal cultures. This conforms to<br />
the findings of a previous investigation (Leuckert et al. 1990).<br />
However, in Graphidaceae little difference between the two<br />
was observed. There were also few differences between the<br />
compounds produced by the fungus cultured under static<br />
conditions compared to those grown in shake culture. In a few<br />
cases, however, some additional compounds were detected<br />
in the shake culture extracts.<br />
Our results, and those of Crittenden et al. (1995) in<br />
particular, demonstrate that, contrary to a general belief<br />
of recalcitrance to grow on artificial media, it is possible<br />
to obtain many lichen-forming fungi in isolated culture –<br />
provided that recently collected material is used. Further,<br />
our results on ascospore discharge show that the seasonal<br />
behaviour and discharge distances of the ascospores of<br />
tropical lichens recalls that of those in temperate regions. We<br />
also suspect that the short distances over which ascospores<br />
are discharged, especially where these are multicelled and<br />
large, contributes to the inability of many to spread into<br />
secondary environments from old-growth native forests and<br />
so facilitates their utility as bioindicators of ecological stability<br />
(Wolesley et al. 1994).<br />
We hope that this preliminary study will encourage more<br />
experimental work on the factors affecting the reproductive<br />
biology of tropical lichens, which are crucial to an<br />
understanding of their ecology and distribution – especially<br />
at local scales.<br />
ACKNOWLEDGEMENTS<br />
We thank the Royal Forest Department of Thailand for permission<br />
to collect samples and for provision of facilities during the surveys<br />
in the national parks of Thailand. We also thank Kansri Boonpragob<br />
(Ramkhamhaeng University), Prakitsin Sihanonth (Chulalongkorn<br />
University), and George P. Sharples (Liverpool John Moores<br />
University) for their valuable comments and laboratory support. This<br />
manuscript was completed while D.L.H. was in receipt of a research<br />
grant from the Ministerio de Ciencia e Innovación in Spain (project<br />
CGL 2008-01600).<br />
References<br />
Ahmadjian V (1964) Further studies on lichenized fungi. Bryologist<br />
67: 87–98.<br />
Ahmadjian V, Jacobs, JB (1981) Relationship between fungus and<br />
alga in the lichen Cladonia cristatella Tuck. Nature 289: 169–172.<br />
152 ima fUNGUS
Tropical lichen fungi: ascospore discharge and culture<br />
Ahmadjian V, Russell LA, Hildreth KC (1980) Artificial reestablishment<br />
of lichens I. morphological interactions between the phycobionts<br />
of different lichens and the mycobionts Cladonia cristatella and<br />
Lecanora chysoleuca. Mycologia 72: 73–89.<br />
Aptroot A, Saipunkaew W, Sipman HJM, Sparrius LB, Wolseley<br />
PA (2007) New lichens from Thailand, mainly microlichens<br />
from Chiang Mai. Fungal Diversity 24: 75–134.<br />
Bailey RH, Garrett RM (1968) Studies on the discharge of ascospores<br />
from lichen apothecia. Lichenologist 4: 57–65.<br />
Bubrick P (1988) Methods for cultivating lichens and isolated bionts.<br />
In: CRC Handbook of Lichenology (M.Galun, ed.) 3: 127–138.<br />
Boca Raton: CRC Press.<br />
Booth C (1971) Fungal culture media. In: Methods in Microbiology<br />
(C. Booth, ed.) 4: 49–94. London: Academic Press.<br />
Crittenden PD, David JC, Hawksworth DL, Campbell FS (1995)<br />
Attempted isolation and success in the culturing of a broad<br />
spectrum of lichen-forming and lichenicolous fungi. New<br />
Phytologist 130: 267–297.<br />
Culberson CF, Armaleo D (1992) Induction of a complete secondaryproduct<br />
pathway in a cultured lichen fungus. Experimental<br />
Mycology 16: 52–63.<br />
Hamada N (1989) The effect of various culture conditions on depside<br />
production by an isolated lichen mycobiont. Bryologist 92: 310–<br />
313.<br />
Homchantara N, Coppins BJ (2002) New species of the lichen family<br />
Thelotremataceae in SE Asia. Lichenologist 34: 113–140.<br />
Honegger R (1990) Mycobiont-photobiont interactions in adult<br />
thalli in axenically resynthesized pre-thallus stage of Xanthoria<br />
parietina (Teloschistales, lichenized ascomycetes). Bibliotheca<br />
Lichenologica 38: 191–208.<br />
Kaye GW, Laby TH (1966) Tables of Physical and Chemical<br />
Constants, and some mathematical functions. 13 th edn. London:<br />
Longmans.<br />
Leuckert C, Ahmadjian V, Culberson CF, Johnson A (1990) Xanthones<br />
and depsidones of the lichen Lecanora dispersa in nature and of<br />
its mycobiont in culture. Mycologia 82: 370–378.<br />
Lumbsch HT, Hunhdorf SB (2010) Myconet Vol. 14. Part One. Outline<br />
of the Ascomycota – 2009. Fieldiana, Life and Earth Sciences 1:<br />
1–40.<br />
Orange A, James PW, White FJ (2001) Microchemical Methods for<br />
the Identification of Lichens. London: British Lichen Society.<br />
Papong K, Boonpragob K, Mangold A, Divakar PK, Lumbsch HT<br />
(2010) Thelotremoid lichen species recently described from<br />
Thailand: a re-evaluation. Lichenologist 42: 131–137.<br />
Sangvichien E (2005) Studies on tropical lichen mycobionts. PhD<br />
thesis, Liverpool John Moores University.<br />
Sipman H, Aptroot A (2001) Where are the missing lichens?<br />
Mycological Research 105: 1433–1439.<br />
Stocker-Wörgötter E, Hager A (2008) Culture methods for lichens<br />
and lichen symbionts. In: Lichen Biology (T.H. Nash iii, ed.): 353-<br />
363. 2 nd edn. Cambridge: Cambridge University Press.<br />
Thomas EA (1939) Über die Biologie von Flechtenbildnern. Beiträge<br />
zur Kryptogamenflora der Schweiz 9: 1–208.<br />
Töbler F (1909) Das physiologische Gleichgewicht von Pilz und<br />
Alge in den Flechten. Berichte der Deutschen Botanischen<br />
Gesellschaft 27: 421–427.<br />
Turbin L (1996) The growth and physiology of lichen-forming fungi.<br />
PhD thesis, University of Nottingham.<br />
Werner R-G (1927) Recherches biologiques et expérmentales sur<br />
les ascomycetes de lichens. Thesis, Docteur des Sciences<br />
Naturelles, Paris.<br />
Wolseley PA, Moncrieff C, Aguirre-Hudson B (1994) Lichens as<br />
indicators of environmental stability and change in the tropical<br />
forests of northern Thailand. Global Ecology and Biogeography<br />
Letters 4: 116–123.<br />
Yamamoto Y, Kinoshita Y, Fujita M (1998) Do lichen mycobionts grow<br />
in liquid culture? Lichen, News Bulletin of the Lichenological<br />
Society of Japan 11(1): 6–7.<br />
Yoshimura I, Yamamoto Y, Nakano T, Finnie J (2002) Isolation and<br />
culture of lichen photobionts and mycobionts. In: Protocols in<br />
Lichenology: culturing, biochemistry, ecophysiology and use in<br />
biomonitoring (Kranner I, Beckett RP, Varma AK, eds): 3–33.<br />
Berlin: Springer-Verlag.<br />
ARTICLE<br />
volume 2 · no. 2<br />
153
ARTICLE<br />
<br />
ima fUNGUS
doi:10.5598/imafungus.2011.02.02.06 <strong>IMA</strong> <strong>Fungus</strong> · volume 2 · no 2: 155–162<br />
A new dawn for the naming of fungi: impacts of decisions made in Melbourne<br />
in July 2011 on the future publication and regulation of fungal names 1<br />
David L. Hawksworth<br />
Departamento de Biología Vegetal II, Facultad de Farmacia, Universidad Complutense de Madrid, Plaza Ramón y Cajal, Madrid 28040, Spain;<br />
and Department of Botany, Natural History Museum, Cromwell Road, London SW7 5BD, UK; corresponding author e-mail: d.hawksworth@nhm.<br />
ac.uk<br />
ARTICLE<br />
Abstract: A personal synopsis of the decisions made at the Nomenclature Section meeting of the<br />
International Botanical Congress in Melbourne in July 2011 is provided, with an emphasis on those which<br />
will affect the working practices of, or will otherwise be of interest to, mycologists. The topics covered include<br />
the re-naming of the Code, the acceptance of English as an alternative to Latin for validating diagnoses,<br />
conditions for permitting electronic publication of names, mandatory deposit of key nomenclatural information<br />
in a recognized repository for the valid publication of fungal names, the discontinuance of dual nomenclature<br />
for pleomorphic fungi, clarification of the typification of sanctioned names, and acceptability of names<br />
originally published under the zoological code. Collectively, these changes are the most fundamental to have<br />
been enacted at a single Congress since the 1950s, and herald the dawn of a new era in the practice of fungal<br />
nomenclature.<br />
Key words:<br />
Amsterdam Declaration<br />
Code of Nomenclature<br />
electronic publication<br />
MycoBank<br />
nomenclature<br />
pleomorphic fungi<br />
registration<br />
sanctioned names<br />
taxonomy<br />
Article info: Submitted 12 September 2011; Accepted 20 September 2011; Published 11 November 2011.<br />
Introduction<br />
The internationally agreed rules that regulate how fungi<br />
are named are examined and revised at each International<br />
Botanical Congress, the last published being those resulting<br />
from the Vienna Congress in 2005 (McNeill et al. 2006).<br />
These Congresses are now held every six years, and the<br />
subsequent one in Melbourne in July 2011 was faced with a<br />
staggering 338 proposals made to modify the Vienna edition<br />
of the International Code of Botanical Nomenclature (McNeill<br />
& Turland 2011). This was the largest number to have<br />
confronted any Congress since that held in Paris in 1954.<br />
The <strong>issue</strong>s that the Melbourne Congress had to address<br />
included topics as fundamental as the language required for<br />
the valid publication of names, the acceptability of electronic<br />
publication, and the unease amongst mycologists on how<br />
decisions were made.<br />
It may seem weird to 21 st century biological science<br />
students that fungi are embraced in a Code with just<br />
“botanical” in the title. However, the actual remit was all<br />
organisms traditionally studied in departments of botany<br />
in museums and universities, regardless of their current<br />
classification in the kingdoms of Life – even all bacteria<br />
were covered until the Montreal Congress of 1959. Some<br />
rules are, nevertheless, applicable only to particular<br />
systematic groups or categories, and since the Brussels<br />
Congress of 1910 there have been special regulations<br />
which only apply to the names of fungi. Foremost amongst<br />
these have been <strong>issue</strong>s related to: (1) the date at which<br />
the nomenclature of fungi was deemed to commence; (2)<br />
the status of living cultures as name-bearing types; and (3)<br />
the separate naming of morphs in pleomorphic fungi. Any<br />
proposed changes in the rules relating to particular groups<br />
or categories (e.g. fossils) are discussed by a series of<br />
permanent committees, the members of which are elected<br />
at the end of each Congress and serve to the next. In the<br />
case of the fungi, the permanent committee is now called<br />
the Nomenclature Committee for Fungi (NCF). A valuable<br />
synopsis of how the current system operates is given by<br />
McNeill & Greuter (1986), while Nicolson (1991) provides<br />
an authoritative historical account of the development of the<br />
Code.<br />
During recent decades, and especially in the 2000s,<br />
many mycologists had become increasingly dissatisfied<br />
with various aspects of the rules concerning the naming<br />
of fungi. This was reflected in sessions and debates at<br />
various national, regional, and international meetings,<br />
culminating in three Nomenclature Sessions held as a part<br />
of the IXth International Mycological Congress (IMC9) in<br />
Edinburgh in August 2010. During those sessions, various<br />
1<br />
This article was first published in MycoKeys 1: 7–20 (2011), doi:<br />
10.3897/mycokeys.1.2062, and is reproduced here with minor<br />
corrections and with the permission of Pensoft Publishers.<br />
© 2011 International Mycological Association<br />
You are free to share - to copy, distribute and transmit the work, under the following conditions:<br />
Attribution:<br />
You must attribute the work in the manner specified by the author or licensor (but not in any way that suggests that they endorse you or your use of the work).<br />
Non-commercial: You may not use this work for commercial purposes.<br />
No derivative works: You may not alter, transform, or build upon this work.<br />
For any reuse or distribution, you must make clear to others the license terms of this work, which can be found at http://creativecommons.org/licenses/by-nc-nd/3.0/legalcode. Any of the above conditions can be waived if you get<br />
permission from the copyright holder. Nothing in this license impairs or restricts the author’s moral rights.<br />
volume 2 · no. 2 155
Hawksworth<br />
ARTICLE<br />
already published proposals for change were discussed,<br />
and in addition all delegates to the Congress were invited<br />
to complete a questionnaire to canvass their views on key<br />
<strong>issue</strong>s and possible ways forward; a report of those Sessions<br />
and the results of the questionnaires are provided by Norvell<br />
et al. (2010).<br />
The decisions taken at the Melbourne Congress<br />
were so fundamental, with respect to both “botanical”<br />
nomenclature as a whole, and especially with specific<br />
topics that concerned fungi, that these need to be widely<br />
promulgated. A formal report of those decisions is provided<br />
by McNeill et al. (2011), and more detailed information of<br />
those pertaining to fungi is presented by Norvell (2011).<br />
Those reports include the new approved wordings, though<br />
they may still undergo some fine-tuning by the Editorial<br />
Committee appointed by the Congress. The Editorial<br />
Committee is to meet in London in December 2011, and it is<br />
anticipated that the finalized Melbourne Code will be printed<br />
in mid-2012. However, changes effected at an International<br />
Botanical Congress come into effect immediately they are<br />
approved by the Plenary Session of the Congress unless<br />
specifically limited by date. It is, therefore, essential that all<br />
mycologists involved in the naming of fungi are made aware<br />
of both the changes made that come into force before the<br />
Code is printed, and those that are to be anticipated from 1<br />
January 2013.<br />
The purpose of the present article is to alert mycologists as<br />
a whole to the fundamental changes made at the Melbourne<br />
Congress, a package which represents a paradigm shift in how<br />
fungi are now to be named, and to indicate the implications<br />
of those changes for working practices. It is not, however,<br />
to be considered authoritative, and the final version of the<br />
Melbourne Code should be consulted as soon as it becomes<br />
available.<br />
Principle changes and their impacts<br />
Name of the Code changed<br />
Mycologists, tired of appearing subservient to botanists, and<br />
for mycology to be treated as a part of botany (Hawksworth<br />
1997, Minter 2011), made proposals for the name of the Code<br />
to be changed to reflect their independence (Hawksworth et<br />
al. 2009). This view had been supported at IMC9 (Norvell<br />
et al. 2010), and the Melbourne Congress agreed that<br />
the new Code should be called the International Code of<br />
Nomenclature for algae, fungi, and plants. The lower case<br />
letters used for the words “algae”, “fungi”, and “plants” are<br />
employed to make clear these terms are being used in a<br />
colloquial sense, for instance the inclusion of cyanobacteria<br />
in algae, and chromistan fungal analogues and slime moulds<br />
in “fungi”.<br />
The Congress further agreed that editorial changes should<br />
be made throughout the text so that it referred to “organisms”<br />
governed by the Code, and no longer used “plants” where<br />
fungi were included in the concept.<br />
Governance of fungal nomenclature to be<br />
considered<br />
Proposals to transfer decision-making on <strong>issue</strong>s concerning<br />
fungi from International Botanical to International Mycological<br />
Congresses (Hawksworth et al. 2009), and which had been<br />
strongly supported at IMC9 (Norvell et al. 2009) were not<br />
accepted. However, a Subcommittee on governance of the<br />
Code with respect to fungi was established under a Special<br />
Committee mandated with examining how the Nomenclature<br />
Section operated. That Committee (and Subcommittee)<br />
are to report to the next International Botanical Congress<br />
in 2017. In view of this move, mycologists will now have to<br />
consider whether to put on hold the question of the need<br />
for an independent Code for fungi (see below) pending that<br />
report. The matter needs to be placed on the agenda for<br />
Nomenclature Sessions to be convened during IMC10 in<br />
2014.<br />
English or Latin validating diagnoses permitted<br />
The <strong>issue</strong> of whether to discontinue the requirement for<br />
validating diagnoses or descriptions in Latin has been raised<br />
at almost all International Botanical Congresses since this<br />
requirement was first introduced in 1935. The Melbourne<br />
Congress was presented with proposals from botanists to<br />
allow any language, as is the practice in zoology, and some<br />
alternative ones, including one by mycologists to require<br />
Latin or English for fungi (Norvell et al. 2010, Demoulin<br />
2010). There was a precedent in that the alternative of Latin<br />
or English was already allowed for fossils in the Vienna Code.<br />
The Congress not only supported the mycological proposal,<br />
but also decided that it should apply not just to fungi but to all<br />
organisms treated under the Code. Further, so enthusiastic<br />
was the meeting, that it was agreed that this provision<br />
should operate from 1 January 2012, not 1 January 2013.<br />
Consequently, mycologists no longer need to struggle with<br />
coining a few sentences of pseudo-Latin when describing<br />
new fungi. However, in consequence, I personally see<br />
value in presenting both a diagnosis (i.e. a short statement<br />
of how the fungus differs from others) and a separate<br />
description (i.e. a detailed account of all the features of the<br />
fungus) when describing a new fungus. If a diagnosis were<br />
in Latin or English, the description could then continue to<br />
be in any language of the author’s choice. A diagnosis has<br />
been required for the introduction of new scientific names in<br />
zoology since 1930 (International Commission on Zoological<br />
Nomenclature 1999: Art. 13), and the practice has much to<br />
commend it.<br />
Electronic publication permitted (but with<br />
restrictions)<br />
The <strong>issue</strong> of the acceptability of works published only<br />
electronically as a vehicle for the effective publication of<br />
scientific names has been the subject of a series of Special<br />
Committees established by successive International Botanical<br />
Congresses since that held in Tokyo in 1993, and is also an<br />
<strong>issue</strong> currently being actively debated by zoologists (Michel et<br />
al. 2009). With the increasing proliferation of new electronic<br />
156 ima fUNGUS
A new dawn for the naming of fungi<br />
journals, and established journals also increasingly being<br />
available in both electronic and hard-copy forms, the <strong>issue</strong><br />
was becoming increasingly urgent. A Special Committee<br />
established by the Vienna Congress in 2005, considered the<br />
matter in depth (Chapman et al. 2010) and prepared detailed<br />
proposals for consideration by the Melbourne Congress<br />
(Special Committee on Electronic Publication 2010). The<br />
Melbourne Congress accepted many of these proposals, and<br />
the pertinent revised texts and guidelines as to best practice<br />
are given by Knapp et al. (2011). The key points agreed were<br />
that from 1 January 2012, works published in electronic form<br />
on the worldwide web in an unchangeable Portable Document<br />
Format (PDF) are to be treated as effectively published,<br />
provided that they have either an International Standard Serial<br />
Number (ISSN) or an International Standard Book Number<br />
(ISBN). However, non-final versions made available online<br />
in advance of a definitive version (e.g. accepted papers as<br />
yet unedited or proof-read) are not treated as effectively<br />
published. Where both electronic and hard-copy versions of<br />
a work are made available, the date of effective publication of<br />
both is treated as being the same. Guidance as to how copies<br />
can be differentiated is included in Knapp et al. (2011).<br />
It is important to appreciate that the new provisions do not<br />
mean that material placed on or available through websites<br />
and lacking ISSN or ISBN numbers constitutes effective<br />
publication. Authors considering submitting to an electronic<br />
journal, should therefore first check that it has an ISSN<br />
number. It is also recommended that electronic-only works<br />
containing new taxa are drawn to the attention of appropriate<br />
indexing centres, and mycologists should endeavour to do that<br />
until the requirement for the prior deposit of key nomenclatural<br />
information becomes mandatory on 1 January 2013.<br />
Deposit of key nomenclatural information<br />
made mandatory for fungi<br />
The concept of some form of obligatory registration of newly<br />
proposed scientific names for fungi goes back to the 1950s<br />
(Ainsworth & Ciferri 1955). Following the establishment of a<br />
Special Committee on Registration at the Berlin Congress in<br />
1987, and a series of subsequent workshops, a provision to<br />
make this a requirement for all groups of organisms covered<br />
by the Code was accepted by the Tokyo Congress in 1993 –<br />
but then rejected at the St Louis Congress in 1999 despite<br />
successful trials (Greuter 2009). The development of the<br />
worldwide web, however, has made it possible to devise<br />
much-improved systems from those that were possible in<br />
the 1980s and early 1990s. Following informal discussions<br />
during the 2002 International Mycological Congress (IMC7)<br />
in Oslo, in 2004 the CBS-KNAW Fungal Biodiversity Centre<br />
in Utrecht established an online system for the deposit of key<br />
information on newly proposed names of fungi – MycoBank.<br />
This voluntary system proved popular with mycologists,<br />
and also with mycological journals, as a way of rapidly<br />
expediting information on nomenclatural novelties. Since<br />
2007 Mycobank has operated under the auspices of the<br />
International Mycological Association (<strong>IMA</strong>) which now has<br />
long-term responsibility for its continuance.<br />
Formal proposals to make the deposit of key nomenclatural<br />
information in a recognized online repository a mandatory<br />
requirement for valid publication of new scientific names in<br />
fungi at all taxonomic ranks (including new combinations and<br />
replacement names) were then developed (Hawksworth et<br />
al. 2010). Those proposals were overwhelming endorsed<br />
by the International Mycological Congress in Edinburgh<br />
later in the same year (Norvell et al. 2010). The Melbourne<br />
Congress approved the formal proposals with some “friendly”<br />
amendments, mainly based on suggestions for avoiding<br />
unnecessary inflation of names in the repositories (Morris<br />
et al. 2011). In addition a recommendation that information<br />
on choices made between competing names or homonyms,<br />
spelling or gender also be deposited (Gams 2010) was<br />
approved.<br />
The new requirement comes into force on 1 January 2013,<br />
after which date scientific names of fungi which are published<br />
without a unique identifier by a recognized repository will not<br />
be considered as validly published; i.e. they will not exist for<br />
nomenclatural purposes and need not be considered when<br />
determining the correct name for a taxon under the Code.<br />
While the requirement is only for information required by the<br />
rules of the Code, such as the diagnosis and information as<br />
to the nomenclatural type or a basionym, as appropriate,<br />
there is no objection to databases also including additional<br />
information and the prospects are enormously exciting<br />
(Lumbsch et al. 2011).<br />
The responsibility of appointing online depositories was<br />
given to the Nomenclature Committee for Fungi, which will<br />
need to advise mycologists as to which are approved. No<br />
single repository was specified in the proposals, thus leaving<br />
the possibilities open in the rapidly-moving electronic age. At<br />
present it is deposit in MycoBank which is now required by<br />
almost all mycological journals.<br />
Mycologists should note that the prudent way to proceed<br />
is to make the online deposit of the required data, and<br />
obtain the numerical identifier, only after their work has been<br />
accepted for publication. This is to ensure that the information<br />
included agrees in every detail that which will appear in the<br />
publication which establishes the name. This will not affect<br />
the priority of the name as the effective date of publication<br />
will be that of the electronic or hard-copy publication and not<br />
the date information is deposited. The lodging of a name and<br />
associated details in a repository such as MycoBank will not<br />
in itself establish a name.<br />
This exciting move means that, for the first time ever,<br />
mycologists will have immediate and free online access to<br />
the key nomenclatural and diagnostic information on newly<br />
proposed fungal names. It also means that it is the authors of<br />
new names which will now have the responsibility of ensuring<br />
that names they propose are incorporated into international<br />
indexing repositories.<br />
Dual nomenclature of pleomorphic fungi<br />
discontinued<br />
The concept of permitting separate names for anamorphs<br />
of fungi with a pleomorphic life-cycle has been an <strong>issue</strong> of<br />
ARTICLE<br />
volume 2 · no. 2<br />
157
Hawksworth<br />
ARTICLE<br />
debate since the phenomenon was recognized in the mid-<br />
19 th century. This was even before the first international rules<br />
for “botanical” nomenclature were <strong>issue</strong>d in 1867 (Weresub<br />
& Pirozynski 1979, Taylor 2011). Special provisions are to<br />
be found in the earliest Codes, which were then modified<br />
several times, and often substantially (Weresub & Pirozynski<br />
1979). The rules became increasingly complex, and by the<br />
mid-1970s they were being interpreted in different ways<br />
by different mycologists – even ones working on the same<br />
genus. Following intensive discussions under the auspices<br />
of the International Mycological Association (<strong>IMA</strong>), drastic<br />
changes were made at the Sydney Congress in 1981 to clarify<br />
and simplify the procedures – and the now familiar terms<br />
anamorph, teleomorph, and holomorph entered general use.<br />
An unfortunate effect of the simplification was that many name<br />
changes had to be made as a consequence, including ones<br />
of some well-known and economically important species; at<br />
that date, the conservation of species names was not allowed<br />
under the Code.<br />
Unforeseen in the 1970s, when the 1981 provisions were<br />
crafted, was the impact of molecular systematics. A decade<br />
later, it was starting to become obvious that fungi with no<br />
known sexual stage could confidently be placed in genera<br />
which were typified by species in which the sexual stage<br />
was known (Reynolds & Taylor 1991), and the <strong>issue</strong> of the<br />
abandonment of the dual nomenclatural system was posited<br />
(Reynolds & Taylor 1992). This possibility was debated at<br />
subsequent International Mycological Congresses, and on<br />
other occasions (e.g. Seifert et al. 2000, Seifert 2003), and<br />
the need for change was increasingly recognized. Cannon<br />
& Kirk (2000) regarded deletion as inevitable in the longterm,<br />
and further calls for deleting the provision followed (e.g.<br />
Rossman & Samuels 2005). At the International Botanical<br />
Congress in Vienna in 2005, some minor modifications<br />
were made which allowed anamorph-typified names to be<br />
epitypified by material showing the sexual stage when it was<br />
discovered, and for that name or epithet to continue to be<br />
used where there was no previously sexually-typified name<br />
available.<br />
More importantly, the Vienna Congress established<br />
a Special Committee to investigate the <strong>issue</strong> further, but<br />
unfortunately it was unable to reach a consensus (Redhead<br />
2010). Matters were becoming increasingly desperate as<br />
mycologists using molecular phylogenetic approaches<br />
started to ignore the provisions, or interpret them in different<br />
ways (Rossman & Seifert 2010). The view that emerged<br />
from the International Mycological Congress in Edinburgh<br />
the same year, was that mycologists, as a whole, favoured<br />
gradual progress towards a single nomenclature (Norvell et<br />
al. 2010). In the meantime, various proposals were made<br />
to improve the situation, but the situation was becoming so<br />
complex that few mycologists were likely to take the time to<br />
understand them fully and implement them correctly. In order<br />
to progress the matter, an international symposium was<br />
held in Amsterdam in April 2011, under the auspices of the<br />
International Commission on the Taxonomy of Fungi (ICTF),<br />
to explore ways to obtain a solution. If a solution could not<br />
be reached at the Melbourne Congress, the prospect was<br />
for no substantive change to be made until after the 2017<br />
International Botanical Congress. This situation would then<br />
have become intolerable as mycologists increasingly ignore<br />
the rules.<br />
The Amsterdam symposium prepared a declaration of<br />
principles which, it was hoped, would be accommodated in<br />
any change made to Article 59 (Hawksworth et al. 2011). In<br />
effect these amounted to the ending of dual nomenclature,<br />
but with safeguards to minimize changes in familiar names.<br />
The “Amsterdam Declaration” prompted a critical response<br />
from some other mycologists who perceived difficulties in<br />
aspects of the declaration, and wished to continue allowing<br />
dual nomenclature (Gams et al. 2011). Both these documents<br />
were made available to delegates at the Melbourne Congress.<br />
In order to ensure some resolution of the <strong>issue</strong>, proposals<br />
for three possible options were developed by Redhead, in<br />
consultation with various mycologists, for presentation at the<br />
meeting. Following extensive discussions at the Congress,<br />
the option to discontinue the dual nomenclature system<br />
was approved, but with some safeguards to limit resultant<br />
instability (Norvell 2011, McNeill et al. 2011).<br />
After 1 January 2013, one fungus can only have one<br />
name; the system of permitting separate names to be used<br />
for anamorphs then ends. This means that all legitimate<br />
names proposed for a species, regardless of what stage<br />
they are typified by, can serve as the correct name for that<br />
species. All names now compete on an equal footing for<br />
priority regardless of the stage represented by the namebearing<br />
type. In order not to render names that had been<br />
introduced in the past for separate morphs as illegitimate, it<br />
was agreed that these should not be treated as superfluous<br />
alternative names in the sense of the Code. It was further<br />
decided that anamorph-typified names should not be taken<br />
up to displace widely used teleomorph-typified names until<br />
the case has been considered by the General Committee<br />
established by the Congress 2 .<br />
Recognizing that there were cases in some groups of<br />
fungi where there could be many names that might merit<br />
formal retention or rejection, a new provision was introduced.<br />
It was decided that lists of names can be submitted to the<br />
General Committee and, after due scrutiny, names accepted<br />
on those lists are to be treated as conserved over competing<br />
synonyms (and listed as Appendices to the Code). Lichenforming<br />
fungi (but not lichenicolous fungi) were always<br />
excluded from the provisions permitting dual nomenclature;<br />
the new Code will include a paragraph to make it explicit that<br />
lichen-forming fungi are excluded from the newly accepted<br />
provisions.<br />
Mycologists need now to work to implement this major<br />
change. In cases where a later teleomorph-typified name is<br />
2<br />
The General Committee is elected at each International Botanical<br />
Congress, and is responsible for receiving, considering, and<br />
approving reports from the various permanent nomenclature<br />
committees, such as the Nomenclature Committee for Fungi, for the<br />
period up to the next Congress.<br />
158 ima fUNGUS
A new dawn for the naming of fungi<br />
not widely used, it can be anticipated that mycologists will<br />
now simply adopt the earlier anamorph-typified name. If<br />
others consider a decision inappropriate, a proposal for the<br />
conservation of the teleomorph-typified name over the earlier<br />
anamorph-typified name can be made to the Nomenclature<br />
Committee for Fungi (NCF). Although no detailed arrangements<br />
were made at the Congress, it is anticipated that, where<br />
specialist working groups on particular fungal genera or<br />
families exist, as is the case for subcommissions of the<br />
International Commission on the Taxonomy of Fungi (ICTF),<br />
draft lists of names for possible approval will be prepared<br />
by them. In my personal view, there could also be some<br />
advantage in endeavouring to have one list covering all<br />
potentially affected generic names, if mechanisms to achieve<br />
that could be put in place. In the early part of 2012, the NCF<br />
is to work closely with the ICTF and other groups where they<br />
exist (e.g. within the International Union of Microbiological<br />
Societies, IUMS) to develop processes for the preparation<br />
of lists on particular groups. Draft lists will need to be made<br />
available for comment by mycologists at large (e.g. through<br />
the <strong>IMA</strong> and ICTF web sites), and they will then require<br />
revising in the light of comments received. Lists received by<br />
the NCF would, after due consideration by that Committee,<br />
then be forwarded to the General Committee for approval.<br />
Where mycologists wish still to refer to anamorphs<br />
separately, the new provisions do not prohibit informal<br />
usages, such as “acremonium-state” or “acremonium-like”,<br />
ideally with a small initial letter and normal not italic type as<br />
suggested by Cannon & Kirk (2000). This form of typography<br />
makes clear that the designations are not scientific names<br />
governed by the Code.<br />
Typification of sanctioned names clarified<br />
The dates on which the nomenclature of fungi was deemed<br />
to start were changed from 1801 or 1821 to 1753 by the<br />
International Botanical Congress in Sydney in 1981. This<br />
change was made because the later-starting point system<br />
had come to be interpreted in different ways, and because<br />
of difficulties in ascertaining the first usages of already<br />
proposed names after the proscribed dates (Demoulin et<br />
al. 1981). In order to minimize the resultant name changes,<br />
the concept of “sanctioning” was introduced. Sanctioning<br />
permitted the continued use of names that had been adopted<br />
in the 1801 Synopsis Methodica Fungorum of Persoon, or<br />
the 1821-32 Systema Mycologicum of Fries over names that<br />
otherwise would have to be taken up under the normal rules<br />
of priority, homonymy, etc. However, the wording of the rule<br />
in the Sydney Code was somewhat ambiguous and, although<br />
modified slightly at the Berlin Congress in 1987, it could still<br />
be interpreted as meaning either that the typification of a<br />
sanctioned name should be made only on materials cited in<br />
the sanctioning work, or that it could be based on materials<br />
cited in the original pre-sanctioning place of publication.<br />
Proposals to address this <strong>issue</strong> were published before the<br />
Melbourne Congress (Perry 2010, Redhead et al. 2010), but<br />
there were concerns over these. In consequence, a series of<br />
informal discussions was held in Melbourne, which involved<br />
the proposers and other concerned mycologists. Those<br />
meetings led to the formulation of a series of proposals<br />
which were adopted by the Congress (McNeill et al. 2011,<br />
Norvell 2011). The net effect of the changes made is that a<br />
name that has been sanctioned can now be lectotypified (not<br />
neotypified) by material from among the elements associated<br />
with either the original protologue of the name, the sanctioning<br />
treatment, or both. A further and welcome clarification is that<br />
in cases where in the sanctioning work elements associated<br />
with the original protologue did not include a subsequently<br />
designated type selected for the sanctioned name, the<br />
sanctioning author is considered to have introduced a later<br />
homonym that is to be retained because of its sanctioned<br />
status.<br />
No particular date was mentioned in the adopted<br />
proposals, which means that they became operative when<br />
approved by the Melbourne Congress. They are also<br />
retroactive, and so safeguard many typifications made since<br />
the 1981 Congress which were based on material cited in the<br />
original protologue, or on material of the sanctioning author<br />
where that differed. The adoption of these clarifications is<br />
most welcome as it removes the need for many typifications<br />
made since 1981 to be revisited, something that could<br />
have had unfortunate implications for the stability of many<br />
sanctioned names.<br />
Names of fungi first described as animals are<br />
validly published<br />
The revelation that Microsporidia, a group traditionally studied<br />
by zoologists, belonged to kingdom Fungi posed a threat to<br />
numerous names in use in the phylum. This situation arose<br />
as, while those names had been correctly published and were<br />
available for use under the provisions of the International<br />
Code of Zoological Nomenclature, many did not meet the<br />
requirements of the botanical Code. At the Vienna Congress<br />
in 2005, it was agreed that names within Microsporidia, and<br />
other organisms that had originally been published under<br />
the zoological code, were to be treated as validly published<br />
under the botanical Code. However, in accordance with the<br />
wishes of workers on these fungi, the Melbourne Congress<br />
accepted proposals made by Redhead et al. (2009) that<br />
these organisms should be excluded from governance by the<br />
botanical Code and continue to be covered by the zoological<br />
one, despite their phylogenetic position. It was further agreed<br />
that this principle should be adopted for other groups of<br />
organisms traditionally treated under other codes.<br />
Explicitly indicate the physiological state of<br />
type cultures<br />
A rule in the current Code allows cultures of algae and fungi<br />
to serve as name-bearing types, provided that they are<br />
“preserved in a metabolically inactive state”. In practice, the<br />
physiological state of cultures designated as types is often not<br />
stated by describing authors. In order make this explicit, it is<br />
now recommended that the phrase “permanently preserved<br />
in a metabolically inactive state”, or equivalent, be used when<br />
cultures are designated as types.<br />
ARTICLE<br />
volume 2 · no. 2<br />
159
Hawksworth<br />
ARTICLE<br />
Names based on fossil parts loose special<br />
provisions<br />
In recent years there have been extensive debates in the<br />
palaeobotanical community on how to revise the provisions<br />
relating to the naming of parts of fossil organisms treated<br />
under the Code – and which applied to fungi as well as<br />
plants. Competing sets of proposals were submitted to the<br />
Melbourne Congress. As in the case of ending the separate<br />
naming of anamorphs in pleomorphic fungi, the Congress<br />
decided to abandon the practice of separately naming parts<br />
of fossils. Consequently, names of fossils which prove to be<br />
parts of a single species will now compete with each other<br />
for priority, in the same way as occurs for names not based<br />
on fossils.<br />
The Draft BioCode and MycoCode need to be<br />
revisited<br />
Moves towards increased harmonization between the various<br />
codes of nomenclature were initiated in 1985. However, the<br />
prospect, in the long-term, of having a set of rules governing<br />
the future nomenclature of all organisms was developed<br />
in the early 1990s (Hawksworth 1995). This culminated in<br />
the publication of a Draft BioCode in 1996 which had been<br />
prepared by the IUBS 3 /IUMS International Committee on<br />
Bionomenclature (ICB) 4 . Little progress was made in taking<br />
the initiative further as the mechanisms and resources to<br />
develop the prerequisite lists of names to be considered<br />
available were not forthcoming. The project was subsequently<br />
revived as a scientific programme of IUBS in 2009, and an<br />
updated Draft BioCode was prepared and released for further<br />
discussion in January 2011 (Greuter et al. 2011). That draft<br />
was the subject of a session and debate at Biosystematics<br />
2011 (which incorporated the International Congress of<br />
Systematic and Evolutionary Biology) in Berlin in February<br />
2011. This initiative was mentioned briefly in the final session<br />
of the Nomenclature Section meetings in Melbourne, but was<br />
not considered in any depth. A suggestion that the Section<br />
establish a Special Committee to liaise with those involved in<br />
the revision of the draft was not approved.<br />
The possibility of having an independent code for<br />
mycology was raised and received considerable vocal<br />
support at the International Mycological Congress (IMC8) in<br />
Cairns in 2006. However, the option of renaming and revising<br />
the botanical Code was the one favoured at the subsequent<br />
Congress in Edinburgh in 2010 (Norvell et al. 2010). The<br />
<strong>issue</strong> was also raised at the Amsterdam symposium in April<br />
2011 which was primarily convened to address the <strong>issue</strong><br />
of dual nomenclature. At that symposium it was suggested<br />
that the BioCode model could provide a framework for the<br />
3<br />
The International Union of Biological Sciences, in which the<br />
International Mycological Association represents general mycology.<br />
future regulation of the nomenclature of fungi (Hawksworth et<br />
al. 2011). Key to any movement in this direction, was seen<br />
as the extent to which the botanical Code would change<br />
to meet the needs of mycologists (Taylor 2011). In view of<br />
the major changes made at the Melbourne Congress, the<br />
<strong>issue</strong> of whether an independent MycoCode is really now<br />
required needs to be debated at the International Mycological<br />
Congress (IMC10) in Bangkok in 2014.<br />
Discussion<br />
I have participated in all International Botanical Congresses<br />
since that held in St Petersburg in 1975, and served on the<br />
Editorial Committee of the botanical Code since 1987. The<br />
progress made in adapting the rules to the needs of both<br />
user and practitioner mycologists over that period has been<br />
considerable. These have included, for example, the change<br />
in starting point, the conservation and rejection of species<br />
names, the designation of interpretive types (“epitypes”),<br />
and allowing living metabolically inactive cultures to<br />
be nomenclatural types. The powers of the permanent<br />
Nomenclature Committees have also been enhanced over<br />
the years, so that they can now recommend rejection of any<br />
name whose adoption is regarded as disadvantageous.<br />
Even against this background of increasing adaptation,<br />
the raft of changes effected at the Melbourne Congress in<br />
2011, has to be seen as the dawn of a new era for botanical<br />
and mycological nomenclature, truly bringing it into the<br />
modern age. The decisions made with respect to the name<br />
of the Code, its coverage, electronic publication, and the<br />
requirement for the deposition of key information in a<br />
recognized depositary as a requirement for the publication<br />
of fungal names, place the Melbourne Code ahead of what<br />
zoologists are currently endeavouring to do.<br />
There is still much to be achieved by mycologists, especially<br />
with respect to the implementation of the consequences of<br />
the end of dual nomenclature for pleomorphic fungi, although<br />
the regulatory mechanisms are now in place. A major <strong>issue</strong><br />
that remains is how best to designate taxa only known from<br />
molecular studies of environmental samples, and to consider<br />
whether that requires any changes in the Code (Hawksworth<br />
et al. 2011, Hibbett et al. 2011, Taylor 2011).<br />
Finally, I must stress that the views and interpretations<br />
presented in this overview are personal, and that mycologists<br />
should check the decisions and verify actual wordings agreed<br />
in Melbourne for themselves, especially in the official report<br />
of the Nomenclature Section meetings (McNeill et al. 2011),<br />
and then the edited published version of the International<br />
Code of Nomenclature for algae, fungi, and plants when it<br />
becomes available in mid-2012.<br />
4<br />
The IUBS/IUMS International Committee on Bionomenclature<br />
comprises representatives of the five internationally mandated<br />
organismal codes of nomenclature: botanical, cultivated plant,<br />
prokaryote, viral, and zoological; it was formally established in 1994.<br />
Acknowledgements<br />
My participation in the Melbourne Congress was supported<br />
through a research grant from the Ministerio de Educación<br />
160 ima fUNGUS
A new dawn for the naming of fungi<br />
y Ciencia of Spain (Proyectos I+D CGL 2008-01600), with<br />
a contribution from the International Union of Biological<br />
Sciences (IUBS).<br />
References<br />
Ainsworth GC, Ciferri R (1955) Mycological taxonomic literature and<br />
publications. Taxon 4: 3–6.<br />
Cannon PF, Kirk PM (2000) The philosophies and practicalities of<br />
amalgamating anamorph and teleomorph concepts. Studies in<br />
Mycology 45: 19–25.<br />
Chapman AD, Turland NJ, Watson MF (2010) Report of the Special<br />
Committee on Electronic Publication. Taxon 59: 1853–1862.<br />
Demoulin V (2010) Proposals to amend Articles 15, 36 and 45. Taxon<br />
59: 1611–1612.<br />
Demoulin V, Hawksworth DL, Korf RP, Pouzar Z (1981) A solution to<br />
the starting point problem in the nomenclature of fungi. Taxon<br />
30: 52–63.<br />
Gams W (2010) Proposals to require deposition of information<br />
concerning typification of names of fungal taxa, with an<br />
associated Recommendation. Taxon 59: 1610–1611.<br />
Gams W, Jaklitsch W, Agerer R, Aguirre-Hudson B, Andersen B,<br />
et al. (2011) A critical reponse to the ‘Amsterdam Declaration’.<br />
Mycotaxon 116: 501–513.<br />
Greuter W (2009) Registration of names: the botanical experience.<br />
Bulletin of Zoological Nomenclature 66: 110–114.<br />
Greuter W, Garrity G, Hawksworth DL, Jahn R, Kirk PM, et al. (2011)<br />
Draft BioCode (2011): principles and rules regulating the naming<br />
of organisms. Bionomina 3: 26–44; Taxon 60: 201-212; Bulletin<br />
of Zoological Nomenclature 68: 10–28.<br />
Hawksworth DL (1995) Steps along the road to a harmonized<br />
bionomenclature. Taxon 44: 447-456.<br />
Hawksworth DL (1997) Orphans in “botanical” diversity. Muelleria 10:<br />
111–123.<br />
Hawksworth DL, Cooper JA, Crous PW, Hyde KD, Iturriaga T, et<br />
al. (2010) Proposals to make the pre-publication deposit of<br />
key nomenclatural information in a recognized repository a<br />
requirement for valid publication of organisms treated as fungi<br />
under the Code. Taxon 59: 660-662; Mycotaxon 111: 514-519.<br />
Hawksworth DL, Crous PW, Dianese JC, Gryzenhout M, Norvell<br />
LL, Seifert KA (2009) Proposals to amend the Code to make it<br />
clear that it covers the nomenclature of fungi, and to modify the<br />
governance with respect to names of organisms treated as fungi.<br />
Taxon 58: 658–659; Mycotaxon 108: 1–4.<br />
Hawksworth DL, Crous PW, Redhead SA, Reynolds DR, Samson<br />
RA, et al. (2011) The Amsterdam Declaration on Fungal<br />
Nomenclature. <strong>IMA</strong> <strong>Fungus</strong> 2: 105–112; Mycotaxon 116: 491–<br />
500.<br />
Hibbett DS, Ohman A, Glotzer D, Nuhn M, Kirk PM, Nilsson RH<br />
(2011) Progress in molecular and morphological taxon discovery<br />
of Fungi and options for formal classification of environmental<br />
sequences. Fungal Biology Reviews 25: 38–47.<br />
International Commission on Zoological Nomenclature (1999)<br />
International Code of Zoological Nomenclature. 4 th edn. London:<br />
International Trust for Zoological Nomenclature.<br />
Knapp S, McNeill J, Turland NJ (2011) Changes to publication<br />
requirements made at the XVIII International Botanical Congress<br />
in Melbourne – what does e-publication mean for you? Taxon 60:<br />
1498–1501; Mycotaxon 117: in press; MycoKeys 1: 21–28.<br />
Lumbsch HT, Miller AN, Begerow D, Penev L (2011) MycoKeys, or<br />
why we need a new journal in mycology? MycoKeys 1: 1–5.<br />
McNeill J, Barrie FR, Burdet HM, Demoulin V, Hawksworth DL, et<br />
al. (eds) (2006) International Code of Botanical Nomenclature<br />
(Vienna Code) adopted by the Seventeenth International<br />
Botanical Congress Vienna, Austria, July 2005. [Regnum<br />
Vegetabile no. 146.] Ruggell: A.G. Ganter Verlag.<br />
McNeill J, Greuter W (1986) Botanical nomenclature. In: Biological<br />
Nomenclature Today (Ride WDL, Younés T, eds): 3–25. [IUBS<br />
Monograph no. 2.] Eynsham, Oxford: IRL Press.<br />
McNeill J, Turland NJ (2011) Synopsis of proposals on botanical<br />
nomenclature – Melbourne 2011: a review of the proposals<br />
concerning the International Code of Botanical Nomenclature<br />
submitted to the XVIII International Botanical Congress. Taxon<br />
60: 243–286.<br />
McNeill J, Turland NJ, Monro A, Lepschi BJ (2011) XVIII International<br />
Botanical Congress: preliminary mail vote and report of Congress<br />
action on nomenclature proposals. Taxon 60: 1507–1520.<br />
Michel E, Nikolaeva S, Dale-Skey N, Tracey S (2009) Contributions<br />
to the discussion on electronic publication. Bulletin of Zoological<br />
Nomenclature 66: 4–19.<br />
Minter DW (2011) What every botanist and zoologist should know –<br />
and what every mycologist should be telling them. <strong>IMA</strong> <strong>Fungus</strong><br />
2: (14)–(18).<br />
Morris PL, Macklin JA, Croft J, Nicholson N, Whitehead G (2011)<br />
Letter of concern regarding Props. (117–119) to amend the ICBN<br />
to require pre-publication deposit of nomenclatural information.<br />
Taxon 116: 513–517<br />
Nicolson DH (1991) A history of botanical nomenclature. Annals of<br />
the Missouri Botanical Garden 78: 33–56.<br />
Norvell LL (2011) Fungal nomenclature. 1. Melbourne approves a<br />
new Code. Mycotaxon 116: 481–490.<br />
Norvell LL, Hawksworth DL, Petersen RH, Redhead SA (2010) IMC9<br />
Edinburgh Nomenclature Sessions. Mycotaxon 113: 503–511;<br />
<strong>IMA</strong> <strong>Fungus</strong> 1: 143–147; Taxon 59: 1867–1868.<br />
Perry G (2010) Proposal to amend the wording of Article 7 Example<br />
7. Taxon 59: 1908–1909.<br />
Redhead SA (2010) Report on the Special Committee on the<br />
Nomenclature of Fungi with a Pleomorphic Life Cycle. Taxon 59:<br />
1863–1866.<br />
Redhead SA, Kirk PM, Keeling PJ, Weiss LM (2009) Proposals to<br />
exclude the phylum Microsporidia from the Code. Taxon 58: 669.<br />
Redhead SA, Norvell LL, Pennycook SR (2010) Proposals to amend<br />
articles regulating the typification of names in sanctioning works.<br />
Taxon 59: 1911–1913.<br />
Reynolds DR, Taylor JW (1991) Nucelic acids and nomenclature:<br />
name stability under Article 59. In: Improving the Stability of<br />
Names: needs and options (Hawksworth DL, ed): 171–177.<br />
[Regnum Vegetabile no. 123.] Königstein: Koeltz Scientific<br />
Books.<br />
Reynolds DR, Taylor JW (1992) Article 59: reinterpretation or<br />
revision? Taxon 41: 91–98.<br />
Rossman AY, Samuels GJ (2005) Towards a single scientific name<br />
for species of fungi. Inoculum 56 (3): 3–6.<br />
ARTICLE<br />
volume 2 · no. 2<br />
161
Hawksworth<br />
ARTICLE<br />
Rossman AY, Seifert KA (2010) Preface: phylogenetic revision of<br />
taxonomic concepts in the Hypocreales and other Ascomycota<br />
– a tribute to Gary J. Samuels. Studies in Mycology 68: iv–viii.<br />
Seifert KA (ed) (2003) Has dual nomenclature for fungi run its<br />
course? The Article 59 debate. Mycotaxon 88: 493–508.<br />
Seifert KA, Gams W, Crous PW, Samuels GJ (eds) (2000) Molecules,<br />
morphology and classification: towards monophyletic genera in<br />
the ascomycetes. Studies in Mycology 45: 1–230.<br />
Special Committee on Electronic Publication (2010) Proposals to<br />
permit electronic publications to be effectively published under<br />
specified conditions. Taxon 59: 1907–1908.<br />
Taylor JW (2011) One <strong>Fungus</strong> = One Name: DNA and fungal<br />
nomenclature twenty years after PCR. <strong>IMA</strong> <strong>Fungus</strong> 2: 113–120.<br />
Weresub LK, Pirozynski KA (1979) Pleomorphism of fungi as treated<br />
in the history of mycology and nomenclature. In: The Whole<br />
<strong>Fungus</strong>; the sexual-asexual synthesis (Kendrick B, ed) 1: 17–30.<br />
Ottawa: National Museums of Canada.<br />
162 <br />
ima fUNGUS
doi:10.5598/imafungus.2011.02.02.07 <strong>IMA</strong> <strong>Fungus</strong> · volume 2 · no 2: 163–171<br />
The inclusion of downy mildews in a multi-locus-dataset and its reanalysis<br />
reveals a high degree of paraphyly in Phytophthora<br />
Fabian Runge 1 , Sabine Telle 2,3 , Sebastian Ploch 2,3 , Elizabeth Savory 4 , Brad Day 4 , Rahul Sharma 2,3,5 , and Marco Thines 2,3,5<br />
1<br />
University of Hohenheim, Institute of Botany 210, D-70593 Stuttgart, Germany<br />
2<br />
Biodiversity and Climate Research Centre (BiK-F), Senckenberganlage 25, D-60325 Frankfurt am Main, Germany; corresponding author e-mail:<br />
marco.thines@senckenberg.de<br />
3<br />
Senckenberg Gesellschaft für Naturforschung, Senckenberganlage 25, D-60325 Frankfurt am Main, Germany<br />
4<br />
Michigan State University, Department of Plant Pathology, 104 Center for Integrated Plant Systems, East Lansing, MI 48824, USA<br />
5<br />
Goethe University Frankfurt am Main, Department of Biological Sciences, Institute of Ecology, Evolution and Diversity, Siesmayerstr. 70,<br />
D-60323 Frankfurt (Main), Germany<br />
ARTICLE<br />
Abstract: Pathogens belonging to the Oomycota, a group of heterokont, fungal-like organisms, are amongst the<br />
most notorious pathogens in agriculture. In particular, the obligate biotrophic downy mildews and the hemibiotrophic<br />
members of the genus Phytophthora are responsible for a huge variety of destructive diseases, including sudden<br />
oak death caused by P. ramorum, potato late blight caused by P. infestans, cucurbit downy mildew caused by<br />
Pseudoperonospora cubensis, and grape downy mildew caused by Plasmopara viticola. About 800 species of<br />
downy mildews and roughly 100 species of Phytophthora are currently accepted, and recent studies have revealed<br />
that these groups are closely related. However, the degree to which Phytophthora is paraphyletic and where<br />
exactly the downy mildews insert into this genus in relation to other clades could not be inferred with certainty to<br />
date. Here we present a molecular phylogeny encompassing all clades of Phytophthora as represented in a multilocus<br />
dataset and two representatives of the monophyletic downy mildews from divergent genera. Our results<br />
demonstrate that Phytophthora is at least six times paraphyletic with respect to the downy mildews. The downy<br />
mildew representatives are consistently nested within clade 4 (contains Phytophthora palmivora), which is placed<br />
sister to clade 1 (contains Phytophthora infestans). This finding would either necessitate placing all downy mildews<br />
and Phytopthora species in a single genus, either under the oldest generic name Peronospora or by conservation<br />
the later name Phytophthora, or the description of at least six new genera within Phytophthora. The complications<br />
of both options are discussed, and it is concluded that the latter is preferable, as it warrants fewer name changes<br />
and is more practical.<br />
Key words:<br />
AU test<br />
downy mildews<br />
multigene phylogeny<br />
Peronosporaceae<br />
Phytophthora<br />
taxonomy<br />
Article info: Submitted 21 September 2011; Accepted 20 October 2011; Published 11 November 2011.<br />
Introduction<br />
Oomycetes are a group of organisms that superficially<br />
resemble fungi in their hyphal growth and absorptive way of<br />
nutrition. However, they are not closely related to Mycota,<br />
but belong to a group of heterokont organisms, Straminipila<br />
(Dick 2001), which also includes diatoms and sea-weeds.<br />
Oomycetes have adapted to parasitism of plants at least three<br />
times, once in the Saprolegniales in the genera Aphanomyces<br />
and Pachymetra (Riethmüller et al. 1999, Diéguez-Uribeondo<br />
et al. 2009), and separately in Albuginales and Peronosporales<br />
(Riethmüller et al. 2002, Hudspeth et al. 2003, Thines et al.<br />
2008). While the evolution of obligate biotrophy seems to be<br />
an ancient occurrence for the white blister rusts (Thines &<br />
Kamoun 2010), the downy mildews have more recently arisen<br />
from Phytophthora-like ancestors (Riethmüller et al. 2002,<br />
Göker et al. 2003, 2007, Thines et al. 2008, 2009, Thines<br />
2009). The close relationship of the downy mildews and<br />
Phytophthora revealed by these studies is in contrast to the<br />
widely used taxonomic classifications of Waterhouse (1973)<br />
and Dick (1984, 2001), in which Phytophthora and Pythium<br />
were grouped together in the family Pythiaceae. Although<br />
Cooke et al. (2000) inferred a position of Peronospora sparsa<br />
as a sister group of clade 4 (as defined in that study) based on<br />
ITS sequences alone, no substantial phylogenetic resolution<br />
was present on the phylogenetic backbone, thus failing to<br />
position this group within the genus Phytophthora. Other<br />
studies (including multi-locus studies) that included both downy<br />
mildew and Phytophthora species have so far not resolved the<br />
placement of downy mildews in relation to the different clades<br />
of Phytophthora (Riethmüller et al. 2002, Göker et al. 2007,<br />
Thines et al. 2009, Giresse et al. 2010). Additionally, Thines et<br />
© 2011 International Mycological Association<br />
You are free to share - to copy, distribute and transmit the work, under the following conditions:<br />
Attribution:<br />
You must attribute the work in the manner specified by the author or licensor (but not in any way that suggests that they endorse you or your use of the work).<br />
Non-commercial: You may not use this work for commercial purposes.<br />
No derivative works: You may not alter, transform, or build upon this work.<br />
For any reuse or distribution, you must make clear to others the license terms of this work, which can be found at http://creativecommons.org/licenses/by-nc-nd/3.0/legalcode. Any of the above conditions can be waived if you get<br />
permission from the copyright holder. Nothing in this license impairs or restricts the author’s moral rights.<br />
volume 2 · no. 2 163
Runge et al.<br />
ARTICLE<br />
al. (2009) demonstrated that the support for the sister-group<br />
relationship of Peronospora and clade 4 inferred by Cooke et<br />
al. (2000) could have been the result of an alignment artefact.<br />
Conversely, a recent study by Blair et al. (2008) addressed the<br />
phylogenetic relationships of Phytophthora species with good<br />
resolution, but no downy mildew was included in that study,<br />
leaving their placement to speculation. Downy mildews have<br />
been shown to be a monophyletic assemblage by Göker et<br />
al. (2007). However, Göker & Stamatakis (2006) later (in spite<br />
of being published earlier than Göker et al. 2007) came to the<br />
conclusion that a placement of Phytophthora clade 1 within the<br />
downy mildews would also be possible, although no support<br />
could be obtained for this scenario. The question of which<br />
is the sister clade of the downy mildews, and how this clade<br />
is embedded among the different lineages of Phytophthora<br />
therefore continues to be controversial, but is fundamental for<br />
understanding the evolution of this group of important plant<br />
pathogens, especially with respect to the evolution of biotrophy.<br />
In addition, the taxonomic status of many Phytophthora<br />
species depends on the degree of paraphyly of the genus. At<br />
least with two clades, 9 and 10, Phytophthora is paraphyletic<br />
with respect to downy mildews (Cooke et al. 2000, Göker et al.<br />
2007, Thines et al. 2009), but so far, the degree of paraphyly<br />
of Phytophthora could not be resolved. Therefore, it was<br />
the aim of this study to resolve the phylogenetic placement<br />
of the monophyletic downy mildews (represented by the two<br />
divergent downy mildew genera for which genome data are<br />
currently available) among Phytophthora and to test this<br />
placement statistically, to further clarify the relationships within<br />
this group of important plant pathogens.<br />
Materials and Methods<br />
All sequences of Phytophthora and Pythium were obtained<br />
from the study of Blair et al. (2008) available in the National<br />
Center for Biotechnology Information (NCBI) nucleotide<br />
database, GenBank. The dataset includes sequences of<br />
seven different loci, and all species for which all seven loci<br />
were not available were discarded, except for two Pythium<br />
species for which only six of the seven loci could be obtained.<br />
This resulted in an overall dataset of 121 species sampled.<br />
The sequences of Phytophthora infestans were used to obtain<br />
homologous sequences from the genome of Hyaloperonspora<br />
arabidopsidis from the NCBI database using BLAST (Altschul<br />
et al. 1997) and from the genome of Pseudoperonospora<br />
cubensis (Tian et al. 2011) using the annotated EST<br />
sequence information. Because no sequence information for<br />
the 28S nuclear ribosomal DNA locus of Pseudoperonospora<br />
cubensis could be obtained from the EST library, which was<br />
enriched for protein-coding genes, sequence information was<br />
obtained from the NCBI database, using a sequence from<br />
the study of Riethmüller et al. (2002). GenBank accession<br />
numbers for all sequences included in the analyses are given<br />
in Table S1 (Supplementary Information, online only).<br />
Each of the seven sets of sequences was edited (i.e.<br />
leading and trailing gaps were removed) using the DNASTAR<br />
computer package v. 8 (Lasergene, Madison, WI), and were<br />
aligned separately using MAFFT v. 6.240 (Katoh et al. 2005)<br />
using a webserver interface (http://www.genome.jp/tools/<br />
mafft/). The G-INS-i algorithm was chosen for all alignments.<br />
Subsequently, the aligned sequences were concatenated<br />
for phylogenetic analyses and no further editing was done<br />
on the alignment to ensure reproducibility and to prevent<br />
introduction of bias. After the removal of leading and trailing<br />
gaps 6282 nucleotide sites were included in the phylogenetic<br />
analyses. These comprised seven loci: 1119 bp of the betatubulin<br />
gene, 493 bp of the 60S ribosomal protein L10 gene,<br />
873 bp of the translation elongation factor 1-alpha gene, 720<br />
bp of the 28S nuclear ribosomal DNA gene, 646 bp of the<br />
glyceraldehyde-3-phosphate dehydrogenase gene, 1438 bp<br />
of the heat shock protein 90 gene, and 993 bp of the enolase<br />
gene. The alignment, together with the tree from the Bayesian<br />
Analysis shown in Fig. 1, has been deposited in TreeBASE<br />
(www.treebase.org) under the accession number S11829.<br />
The general time reversible (GTR) model was selected for<br />
the concatenated alignment using Modeltest v. 3.7 (Posada<br />
& Crandall 1998) and PAUP v. 4.0b10 (Swofford 2002), with<br />
gamma-distributed substitution rates (shape parameter =<br />
0.69) and proportion of invariable sites (pinv = 0.54). The<br />
values of these parameters were included in the Bayesian<br />
and Minimum Evolution analyses.<br />
Minimum Evolution (ME) analysis was done using MEGA<br />
v. 4.0 (Tamura et al. 2007), with the gamma-distributed<br />
substitution rates as inferred by Modeltest and using<br />
the Maximum-Composite-Likelihood substitution model.<br />
For inferring tree robustness, 1000 bootstrap replicates<br />
(Felsenstein 1985) were computed.<br />
For Maximum Likelihood (ML) inference, the RAxML<br />
webserver at http://phylobench.vital-it.ch/raxml-bb/<br />
(Stamatakis et al. 2008) was used with standard settings<br />
and maximum likelihood search, including an estimation of<br />
invariable sites. The analysis was repeated five times with<br />
100 bootstrap replicates each. The bootstrap support values<br />
obtained were averaged, because the rapid bootstrapping<br />
algorithm can lead to some deviation.<br />
For Bayesian analysis, MrBayes (Huelsenbeck & Ronquist<br />
2001) at the Phylemon2 webserver (http://phylemon.bioinfo.<br />
cipf.es/) and at a local server, for parallel runs, was used.<br />
Four incrementally heated simultaneous Markov Chain<br />
Monte Carlo chains were run for two million generations with<br />
every 1000 th tree sampled, under the general time reversible<br />
(GTR) model with the gamma-distributed substitution rates<br />
and proportion of invariable sites as inferred by Modeltest.<br />
Maintaining that the standard deviation of split frequencies<br />
was constantly below 0.01 and the stationary phase of the<br />
likelihood values was reached after 10 % of sampled trees<br />
when quitting the analysis. The first 1000 trees sampled this<br />
way were discarded, and the remaining 1000 trees were<br />
used to compute a 50 % majority rule consensus tree and<br />
to estimate the posterior probabilities. To ensure general<br />
reproducibility, the analysis was repeated twice using the<br />
webserver, and twice on a local server using MrBayes v.<br />
3.1.2.<br />
164 ima fUNGUS
High degree of paraphyly in Phytophthora<br />
-/-/0.53<br />
87 4<br />
*<br />
*<br />
9<br />
67/78/*<br />
*<br />
2<br />
5<br />
53/-/0.53<br />
91/97/*<br />
10<br />
*/99/*<br />
57/73/0.99<br />
10<br />
51/61/*<br />
14<br />
64<br />
*<br />
*<br />
84<br />
-<br />
0.66<br />
2<br />
94<br />
*<br />
*<br />
10<br />
97<br />
99<br />
*<br />
21<br />
9<br />
-<br />
86<br />
*<br />
3<br />
-<br />
80<br />
*<br />
2<br />
80/*/* 82<br />
84<br />
22 *<br />
<br />
*<br />
99/99/*<br />
98/*/* Ph. cactorum P0715<br />
77/*/* Ph. cactorum P0714<br />
* Ph. hedraiandra P11056<br />
51 Ph. idaei P6767<br />
Ph. pseudotsugae P10339<br />
93/*/* Ph. clandestina P3942<br />
* Ph. iranica P3882<br />
37<br />
Ph. tentaculata P8497<br />
*<br />
Ph. infestans P10650<br />
93/*/*<br />
Ph. infestans P10651<br />
Ph. andina P13365<br />
Ph. ipomoeae P10227<br />
Ph. ipomoeae P10225<br />
Phytophthora s.str.<br />
* Ph. ipomoeae P10226<br />
*<br />
118 Ph. mirabilis P3005<br />
* Ph. phaseoli P10145<br />
Ph. phaseoli P10150<br />
Ph. nicotianae P10802<br />
Ph. nicotianae P6303<br />
Ph. nicotianae P7146<br />
Ph. nicotianae P10318<br />
* Ph. nicotianae P10116<br />
Ph. nicotianae P1452<br />
Ph. arecae P10213<br />
Ph. palmivora P0255<br />
Ph. palmivora P0113<br />
Ph. quercetorum MD9-2<br />
Ph. megakarya P8516<br />
99/*/*<br />
Pseudoperonospora cubensis<br />
107<br />
Hyaloperonospora arabidopsidis<br />
Ph. quercina P10441<br />
Ph. quercina P10334<br />
Ph. inflata P10341<br />
Ph. citrophthora P6310<br />
*<br />
51 Ph. colocasiae P6317<br />
Ph. botryosa P6945<br />
Ph. meadii P6128<br />
Ph. capsici P1319<br />
90/99/*<br />
Ph. capsici P1314<br />
Ph. capsici P10735<br />
25 97 *<br />
Ph. capsici P0253<br />
* Ph. capsici P10386<br />
* Ph. mexicana P0646<br />
Ph. glovera P10619<br />
* Ph. tropicalis P10329<br />
*<br />
Ph. capsici P10452<br />
12 Ph. capsici P0630<br />
Ph. sp. P10417<br />
Ph. citricola P7902<br />
* Ph. multivesiculata P10327<br />
Ph. multivesiculata P10410<br />
Ph. bisheria P10117<br />
Ph. bisheria P1620<br />
* Ph. katsurae P3389<br />
Ph. katsurae P10187<br />
Ph. heveae P10167<br />
Ph. ilicis P3939<br />
Ph. psychrophila P10433<br />
Ph. nemorosa P10288<br />
Ph. pseudosyringae P10437<br />
Ph. inundata P8478<br />
* Ph. sp. P8619<br />
Ph. humicola P3826<br />
Ph. megasperma P3136<br />
Ph. gonapodyides P10337<br />
* Ph. asparagi P10690<br />
Ph. asparagi P10705<br />
Ph. alni P10568<br />
* Ph. cambivora P0592<br />
Ph. fragariae P3821<br />
* Ph. europaea P10324<br />
28<br />
Ph. uliginosa P10328<br />
* Ph. sinensis P1475<br />
* 94<br />
Ph. melonis P10994<br />
20 86 Ph. cajani P3105<br />
* * Ph. vignae P3019<br />
ê *<br />
Ph. sojae P3114<br />
7 Ph. niederhauserii P10617<br />
*<br />
* Ph. cinnamomi P2159<br />
Ph. cinnamomi P8495<br />
96/99/*<br />
Ph. erythroseptica P1699<br />
Ph. erythroseptica P10385<br />
Ph. erythroseptica P10382<br />
* Ph. richardiae P10355<br />
Ph. richardiae P10811<br />
Ph. richardiae P10359<br />
* Ph. richardiae P7788<br />
Ph. sp. P10672<br />
Ph. cryptogea P1088<br />
Ph. kelmania P10613<br />
99/*/* Ph. drechsleri P10331<br />
46<br />
* Ph. medicaginis P10683<br />
Ph. trifolii P7010<br />
Ph. sansomea P3163<br />
*<br />
-<br />
*<br />
*<br />
4<br />
98<br />
*<br />
-/56/0.93 70<br />
93<br />
* 92<br />
59 8 94<br />
0.91 *<br />
1<br />
7<br />
7<br />
*<br />
*<br />
*<br />
*<br />
35<br />
80<br />
-/*/*<br />
*<br />
88<br />
*<br />
Ph. primulae P10220<br />
* Ph. primulae P10224<br />
Ph. primulae P10333<br />
Ph. porri P6207<br />
*<br />
Ph. porri P10728<br />
Ph. brassicae P10153<br />
*<br />
Ph. brassicae P10414<br />
Ph. brassicae P10154<br />
Ph. syringae P10330<br />
Ph. ramorum P10301<br />
Ph. lateralis P3888<br />
Ph. hibernalis P3822<br />
Ph. foliorum P10969<br />
Ph. macrochlamydospora P10267<br />
* Ph. cuyabensis P8218<br />
Ph. cuyabensis P8224<br />
Ph. lagoariana P8223<br />
Ph. lagoariana P8618<br />
Ph. polonica P15004<br />
* Ph. polonica P15005<br />
Ph. polonica P15001<br />
*<br />
Ph. insolita P6703<br />
Ph. insolita P6195<br />
* Ph. fallax P10725<br />
Ph. captiosa P10719<br />
*<br />
Ph. kernoviae P10681<br />
49<br />
Ph. boehmeriae P6950<br />
Pythium vexans P3980<br />
Pythium undulatum P10342<br />
1a<br />
1b<br />
1c<br />
1.1<br />
4.2<br />
DM<br />
4.1<br />
2a<br />
2b<br />
7a<br />
7b<br />
8a<br />
8b<br />
8c<br />
9.1<br />
9.2<br />
1<br />
4<br />
2<br />
5<br />
3<br />
6<br />
7<br />
8<br />
9<br />
10<br />
ARTICLE<br />
0.02<br />
Fig. 1. Phylogenetic reconstruction for Phytophthora and the downy mildews (Bayesian Analysis), with support values in Minimum Evolution,<br />
Maximum Likelihood, and Bayesian Analysis, in the respective order, on the branches, and Bremer support below the branches. Small Asterisks<br />
denote maximum support in a single analysis, big asterisks denote maximum support in all three phylogenetic analyses. Clade designations are<br />
those of Blair et al. (2008), with some additional differentiation corresponding to the statistical testing of the tree topology as given in Table 1.<br />
Predominantly caducous and papillate clades are highlighted in blue, the clade containing downy mildews is highlighted in green and the clades<br />
with predominantly non-caducous, non-papillate or semi-papillate members are highlighted in brown. For Phytophthora, the highlighted areas<br />
are divided into blocks representing groups that lead to paraphyly of Phytophthora and could potentially serve as a basis for the description of<br />
new genera.<br />
volume 2 · no. 2<br />
165
Runge et al.<br />
ARTICLE<br />
Table 1. Results of the site-wise log-likelihoods generated under possible associations of species in base edges. The first column gives the<br />
possible associations for which the site-wise log-likelihoods were produced. Columns show the support values for the approximately unbiased<br />
(AU) test, the observed log-likelihood differences of the edges (OBS), Bootstrap probability tests (NP, BP; and PP), Kishino-Hasegawa (KH)<br />
test, Shimodaira-Hasegawa (SH) test, weighted Kishino-Hasegawa (WKH) test, and the weighted Shimodaira-Hasegawa (WSH) test.<br />
Possible associations AU OBS NP BP PP KH SH WKH WSH<br />
(4.2, DM) 0,983 -106,9 0,992 0,993 1,000 0,966 0,992 0,974 0,989<br />
(1, 4, DM) 0,983 -106,9 0,992 0,993 1,000 0,966 0,992 0,974 0,989<br />
(1, 2, 4, DM) 0,983 -106,9 0,992 0,993 1,000 0,966 0,992 0,974 0,989<br />
(4, DM) 0,979 -39,4 0,985 0,985 1,000 0,901 0,988 0,94 0,996<br />
(1c,1b) 0,882 -32,7 0,981 0,981 1,000 0,860 0,925 0,925 0,925<br />
(3, 6) 0,713 -28,2 0,918 0,919 1,000 0,753 0,753 0,753 0,753<br />
(1–8, 9.1, DM) 0,679 -14,1 0,648 0,646 1,000 0,721 0,909 0,666 0,916<br />
(1–4, 6, DM) 0,670 -5,6 0,47 0,467 0,997 0,592 0,967 0,592 0,967<br />
(2b, 2.2) 0,644 -5,1 0,407 0,399 0,973 0,593 0,911 0,593 0,927<br />
(5, 7) 0,617 -14,7 0,741 0,742 1,000 0,653 0,807 0,653 0,831<br />
(1, 2, 4, 5, 7, DM) 0,555 5,6 0,104 0,103 0,002 0,408 0,949 0,408 0,951<br />
(1, 2, 4, 5, DM) 0,440 14,7 0,251 0,252 0,000 0,347 0,815 0,347 0,806<br />
(1–6, DM) 0,383 14,7 0,259 0,258 0,000 0,347 0,678 0,347 0,676<br />
(2.1, 2b) 0,356 5,1 0,593 0,601 0,027 0,407 0,585 0,407 0,569<br />
(9.1,9.2) 0,321 14,1 0,352 0,354 0,000 0,279 0,678 0,334 0,668<br />
(3,5–7) 0,302 5,8 0,093 0,091 0,000 0,232 0,911 0,232 0,821<br />
(1–5, 7, DM) 0,287 28,2 0,082 0,081 0,000 0,247 0,636 0,247 0,645<br />
(1–4, DM) 0,287 28,2 0,082 0,081 0,000 0,247 0,636 0,247 0,645<br />
(1b,1.1) 0,118 32,7 0,019 0,019 0,000 0,140 0,596 0,075 0,330<br />
(3, 6, DM) 0,022 106,9 0,007 0,006 0,000 0,034 0,093 0,015 0,065<br />
(1, 4.1) 0,021 39,4 0,015 0,015 0,000 0,099 0,406 0,031 0,156<br />
(1, 4) 0,017 106,9 0,008 0,007 0,000 0,034 0,093 0,015 0,051<br />
(1, 2, 4, 5) 0,017 106,9 0,008 0,007 0,000 0,034 0,093 0,015 0,051<br />
(1, 2, 4) 0,017 106,9 0,008 0,007 0,000 0,034 0,093 0,015 0,051<br />
The following species were randomly chosen as representatives for the corresponding clades and subclades in the statistical analysis –<br />
1c, Phytophthora cactorum ; 1b, P. nicotianae; 1c, P. iranica; 1.1, P. infestans; 2ab, P. capsici; 2.1, P. bisheria; 2.2, P. multivesiculata;<br />
3, P. nemorosa; 4.1, P. quercina; 4.2, P. palmivora; 5, P. katsurae; 6, P. humicola; 7, P. europaea; 8, P. ramorum; 9.1, P. polonica;<br />
9.2, P. captiosa; 10, P. boehmeriae; DM, Pseudoperonospora cubensis.<br />
Inference of Bremer support was done using Maximum<br />
Parsimony with the Parsimony Ratchet implemented in<br />
PRAP2 (Müller 2003), using PAUP v. 4.0b10. The starting<br />
tree was obtained by stepwise addition and subsequently the<br />
tree-bisection-and-reconnection (TBR) algorithm was used.<br />
Two hundred replicates were run with 25 % randomly chosen<br />
characters weighted double and the shortest tree of each<br />
run was saved. Afterwards the decay index of each of the<br />
bisections was obtained in PRAP2.<br />
The Approximately Unbiased (AU) test (Shimodaira 2002)<br />
was applied to the 100 bootstrap replicate trees of the first<br />
Maximum Likelihood analysis and to the last 100 sampled<br />
trees of the first Bayesian Analysis using the CONSEL<br />
computer package (Shimodaira & Hasegawa 2001). The<br />
respectively most probable trees were compared to the<br />
topologies of the resulting trees of the ML, ME and Bayesian<br />
analyses and no conflicting support was found to be present.<br />
For conducting the AU testing of the position of the downy<br />
mildews within Phytophthora and additional statistical tests,<br />
representatives of each of the clades at a node important<br />
to infer the position of the downy mildews or the major<br />
monophyletic clades were chosen. For these 18 accessions,<br />
a Bayesian analysis was conducted as described above, but<br />
with estimation of the gamma-distribution and the proportion<br />
of invariable sites by MrBayes, for enabling the AU testing<br />
with CONSEL. The sampled accessions are given in Table<br />
1. The resulting tree was compared to the original tree<br />
and no conflicting support was present, and only minor<br />
changes in topology (placement of clade 5) were observed,<br />
ensuring the validity of the results. One hundred trees (i.e.<br />
every 20 000 th generation) of the Bayesian analysis were<br />
used to create a site-wise log-likelihood output in PAUP for<br />
bootstrap analysis and statistical testing in CONSEL. The<br />
TREEASS program of the CONSEL computer package<br />
assesses support for each possible association of species<br />
in base edges in the underlying trees and outputs p-values<br />
for the AU test, Bootstrap probability tests (NP, BP; and PP),<br />
Kishino-Hasegawa (KH) test, Shimodaira-Hasegawa (SH)<br />
166 ima fUNGUS
High degree of paraphyly in Phytophthora<br />
test, weighted Kishino-Hasegawa (WKH) test, and weighted<br />
Shimodaira-Hasegawa (WSH) test. Default settings of 10<br />
scaling factors of 0.5–1.4, with 10 000 pseudoreplicates<br />
for each, were used. Phytophthora boehmeria, of the most<br />
basal clade of Phytophthora, was used as an outgroup for<br />
the analyses.<br />
Results<br />
When used independently, the loci of the concatenated<br />
alignment always yielded topologies with no significantly<br />
supported inconsistencies (data not shown). The Maximum<br />
Likelihood (ML) analysis of the concatenated alignment<br />
resulted in a best tree with a log-likelihood of -62481.32, a<br />
Minimum Evolution (ME) tree with a sum of branch lengths of<br />
1.04068070, and the best tree from Bayesian Analysis (BA)<br />
had a log-likelihood score of -62678.74. The best tree from<br />
the BA, with posterior probabilities and bootstrap support<br />
values from the other analyses, is given in Fig. 1. In addition,<br />
Bremer support values are given for all clades and subclades.<br />
Under the given tree, Bremer decay indices > 5 can be<br />
considered as significant support and values of 10 or higher<br />
as strong support. It should be noted that the Bremer support<br />
is not linearly correlated with bootstrap support. Species<br />
of Phytophthora were grouped into nine highly supported<br />
clades, with clade 9 also including clade 10 of Blair et al.<br />
(2008). Tree topology was similar to the one found in Blair et<br />
al. (2008) and no supported conflicts were observed, with the<br />
exception of the before-mentioned inclusion of clade 10 into<br />
clade 9. Downy mildews, represented by the two divergent<br />
genera, Hyaloperonospora and Pseudoperonospora, were<br />
grouped together with maximum support in ML and BA<br />
and strong support in ME inference, and were consistently<br />
found among the members of clade 4 of Blair et al. (2008)<br />
with varying support in the full dataset (Fig. 1). The sistergroup<br />
relationship of downy mildews with a part of clade 4,<br />
comprised of Phytophthora megakarya, P. quercetorum, P.<br />
palmivora, and P. areceae received 70 % bootstrap support<br />
in ME, 59 % in ML and a posterior probability of 0.91, at a<br />
confidence interval at 95 % for the trees sampled. This<br />
group was found sister to P. quercina, although this grouping<br />
received significant support only in the BA. Clade 1 and the<br />
monophyletic group containing the downy mildews and the<br />
clade 4 species of Phytophthora were consistently grouped<br />
together in all analyses, with varying support of 57 % bootstrap<br />
support in ME, 73 % in ML, and a posterior probability of 0.99.<br />
The Bremer decay index was 7 for the grouping of DM with<br />
P. megakarya, P. quercetorum, P. palmivora, and P. areceae<br />
and also 7 for the sister-group placement of the above<br />
assemblage with P. quercina. The sister-group relationship<br />
of clade 1 with clade 4 (including downy mildews) was<br />
supported by a Bremer decay index of 10, thus providing<br />
an independent support for the monophyly of this grouping.<br />
The monophyly of clade 1 was well supported with moderate<br />
to maximum support in the phylogenetic analyses and a<br />
Bremer decay index of 24. The monophyly of clades 2 and<br />
5 was also strongly supported; however, their sister-group<br />
relationship did not receive significant support in any of the<br />
analyses. Clades 1, 4 (plus downy mildews), 2, and 5 were<br />
grouped together with weak support in ME and ML analyses,<br />
but maximum support in the BA. This group was grouped<br />
together with clades 3, 6, and 7 with weak support in ME (67<br />
%), moderate support in ML (78 %) and maximum support in<br />
the BA. Clades 3, 6, and 7 were all found to be monophyletic<br />
with strong to maximum support in all analyses. However,<br />
their grouping as a monophyletic assemblage received only<br />
weak support in ME and BA. Clade 8 was placed basal to<br />
the before-mentioned clades 1–7 and its monophyly received<br />
strong to maximum support in all analyses. A deep divergence<br />
was found between clades 1–8 on the one side and clades<br />
9 and 10 on the other side, resulting in a strong to maximum<br />
support for the monophyly of the assemblage comprised of<br />
clades 1–8 in all phylogenetic analyses, and a Bremer decay<br />
index of 10. Clade 10 was found to be nested within clade<br />
9 in ML and BA, and the monophyly of the group containing<br />
these clades was weakly supported in ME, but strongly<br />
supported in ML and BA, and also received a Bremer decay<br />
index of 9. In the reduced dataset (Fig. S1, Supplementary<br />
Information, online only) the downy mildews, represented<br />
by Pseudoperonospora cubensis, grouped together with<br />
Phytophthora palmivora of clade 4 with maximum support,<br />
and P. quercina was found to be the sister taxon of this<br />
group with strong statistical support. The group comprising<br />
the downy mildew and clade 4 representatives was found<br />
to be sister to clade 1 with maximum support. An alternative<br />
topology was observed for some weakly supported nodes,<br />
as the grouping of clades 3 and 6 as well as the grouping of<br />
clades 5 and 7 received significant support.<br />
To test the robustness of the observed grouping of the<br />
clades, especially with respect to the placement of the downy<br />
mildews within Phytophthora, and to infer the probability of<br />
alternative groupings, several tests were performed, which<br />
are summarised in Table 1. The analyses were carried out<br />
without constraints, seeking for all possible groupings of<br />
the clades and subclades of Phytophthora and the downy<br />
mildews. The clustering of downy mildews with clade 4.2 had<br />
the highest AU values and also received the highest scores in<br />
all other analyses, and also the larger clusters of clades 1, 4,<br />
and DM, and 1, 2, 4, and DM scored equally high. The latter<br />
of these groupings is, in contrast to the tree presented in<br />
Fig. 1, as it excludes clade 5, which was grouped together in<br />
the full phylogenetic analysis with clade 2 without significant<br />
support. But in the phylogeny of the clade representatives,<br />
the grouping that scored high in the AU analysis could also be<br />
observed (Fig. S1). The nesting of the downy mildews within<br />
clade 4 received almost equally high support, with 0.979 in<br />
the AU analysis. Thus the topology of the tree presented<br />
in Fig. 1 with respect to the immediate relationships of the<br />
downy mildews received the highest support in the AU<br />
analysis and all other tests employed. Only four contradicting<br />
clusters were found to be possible. These include an<br />
alternative placement of the downy mildews with clades 3<br />
and 6; the clustering of clades 1 and 4 with the exclusion of<br />
ARTICLE<br />
volume 2 · no. 2<br />
167
Runge et al.<br />
ARTICLE<br />
downy mildews; the clustering of clades 1, 2, 4 and 5 with<br />
the exclusion of downy mildews; and the clustering of clades<br />
1, 2, and 4 with the exclusion of downy mildews. But the<br />
high improbability of these groupings is reflected by very low<br />
AU scores, which were 0.022 for the first and 0.017 for the<br />
other groupings. Groupings of Phytophthora which received<br />
significant support are the clustering of clades 1b and 1c (AU<br />
0.882); although these scored less than for the position of<br />
downy mildews as a sister group of clade 4.2 and their nested<br />
placement in clade 4. The grouping of clades 3 and 6, which<br />
were affiliated to other clades without significant support in<br />
the phylogenetic analyses, received moderate support (AU<br />
0.713). Another grouping which was not observed in the<br />
phylogenetic analysis is the clustering of clades 5 and 7,<br />
which was also moderately supported (AU 0.617). Moderate<br />
support was also obtained for the grouping of clades 1–8,<br />
including downy mildews, together with 9.1 (AU 0.679), and<br />
clades 1–4, including downy mildews, together with clade 6<br />
(AU 0.670).<br />
Discussion<br />
The genus Phytophthora is one of the largest genera of<br />
the oomycetes and contains about 100 currently accepted<br />
species, of which about 60 species were included in the<br />
monograph of Erwin & Ribeiro (1996), and to which about<br />
40 species have been added subsequently (Érsek & Ribeiro<br />
2010). As many of the species are of ecological and economic<br />
interest, Phytophthora has received much attention in the past<br />
decades, and as a consequence, the genome sequencing<br />
of several of its members has been undertaken (Tyler et al.<br />
2006, Haas et al. 2009). New species are being discovered in<br />
the previously species-poor basal clades (Brasier et al. 2005,<br />
Belbahri et al. 2006, Dick et al. 2006), and it seems likely<br />
that only a small fraction of the evolutionary diversity of this<br />
genus has been discovered. The genus Phytophthora has<br />
often been considered a member of Pythiaceae (Waterhouse<br />
1973, Dick et al. 1984, Dick 2001), while the obligate<br />
biotrophic downy mildews were viewed as constituting the<br />
family Peronosporaceae. Dick et al. (1984) even placed the<br />
Peronosporaceae together with the Albuginaceae into the<br />
order Peronosporales and opposed this to the cultivable<br />
Pythiales, which also included Phytophthora. However,<br />
Gäumann (1952) already realised that Phytophthora and<br />
the downy mildews were likely to be closely related, and this<br />
hypothesis was later corroborated with the first molecular<br />
phylogenies including members of both Phytophthora and the<br />
downy mildews (Cooke et al. 2000, Riethmüller et al. 2002).<br />
The strict split between downy mildews and Phytophthora<br />
is rather synthetic, as there are species with intermediate<br />
character states that bridge the apparent gulf between the<br />
necrotrophic and hemibiotrophic members of Phytophthora<br />
and the obligate biotrophic downy mildews (Thines 2009).<br />
For example, the downy mildew genus Viennotia (Göker et<br />
al. 2003) possesses sporangiophores capable of additional<br />
growth after sporulation, Poakatesthia (Thines et al. 2007)<br />
forms intracellular mycelium apart from haustoria, and<br />
Sclerophthora has hyphal sporangiophores which do not<br />
form sporangia simultaneously (Payak & Renfro 1967). All of<br />
these features are usually attributed to Phytophthora species,<br />
although other characteristics place these genera among the<br />
downy mildews (Thines 2009). The chimeric appearance of<br />
Sclerophthora is so pronounced that it was even included in<br />
the monograph of Phytophthora by Erwin & Ribeiro (1996). It<br />
is also noteworthy that evolution of the downy mildews may<br />
have been initiated as parasites of grass relatives (Thines<br />
et al. 2007, Thines 2009). Support for this hypothesis is<br />
provided by Phytophthora species from Cyperaceae which<br />
have also been considered members of an independent<br />
genus, Kawakamia, and are not readily cultivable (Erwin &<br />
Ribeiro 1996). On the other hand, there are reports of axenic<br />
cultivation for Sclerospora graminicola (Tiwari & Arya 1969)<br />
and Sclerophthora macrospora (Tokura 1975), although these<br />
results have not been confirmed by independent experiments<br />
of other groups. Unfortunately, none of the above-mentioned<br />
parasites of grasses could be included in the present study<br />
because of difficulties of amplification using the primers<br />
available. Also, for downy mildews in general, the primers<br />
used by Blair et al. (2008) do not readily amplify the targeted<br />
genes, therefore we obtained these sequences directly from<br />
the genomes of Hyaloperonospora arabidopsidis (Baxter<br />
et al. 2010) and Pseudoperonospora cubensis (Tian et al.<br />
2011). However, as the downy mildews most likely represent<br />
a monophyletic group (Göker et al. 2007), the inclusion of only<br />
these two exemplars from largely divergent downy mildew<br />
genera can be considered valid for inferring the placement<br />
of this group amongst the phylogenetic lineages currently<br />
placed in Phytophthora.<br />
The topology of the tree shown here is mostly congruent<br />
with the topology presented by Blair et al. (2008). However,<br />
the inclusion of the downy mildews has in some cases resulted<br />
in lower support values, especially on the backbone and to<br />
a grouping of clades 2 and 5 without significant support. In<br />
Blair et al. (2008), clade 5 was inferred as being basal to<br />
clade 2 with weak to moderate support. In our investigations,<br />
however, the downy mildews were consistently grouped<br />
together with some members of clade 4, which is in line with<br />
the sister-group relationship for Peronospora sparsa with a<br />
group made up of Phytophthora arecae, P. palmivora, and<br />
P. megakarya as observed by Cooke et al. (2000) on the<br />
basis of ITS sequence data, although it cannot be ruled out<br />
that the finding in that study was influenced by alignment<br />
artefacts (Thines et al. 2009) and a bias of the Neighbourjoining<br />
analysis. In our study, which is based on the multilocus<br />
dataset of Blair et al. (2008) to which sequences from<br />
downy mildew representatives have been added, the close<br />
relationship of the downy mildews with members of clade<br />
4 is also supported by several phylogenetic methods and<br />
statistical tests, in which the sister-group relationship of<br />
clade 4.2 with the downy mildews and the grouping of downy<br />
mildews within clade 4 as a whole received strong support. As<br />
discussed in previous publications on the global phylogeny of<br />
Phytophthora (e.g. Blair et al. 2008, Cooke et al. 2000, Kroon<br />
168 ima fUNGUS
High degree of paraphyly in Phytophthora<br />
et al. 2004), there are no clear-cut synapomorphies identified<br />
for the different clades so far. However, four of the five groups<br />
with predominantly papillate or caducous sporangia (1, 2, 4,<br />
and 5), together with the downy mildews, form the crown<br />
group of Phytophthora, and it is thus likely that caducous<br />
and papillate sporangia represent a derived character state.<br />
This is in contrast to the conclusion of Kroon et al. (2004),<br />
who, based on a smaller set of loci, deduced that papillate<br />
sporangia could also be a plesiomorphic trait. Clade 3, which<br />
was considered papillate by Kroon et al. (2003), was found<br />
to sister to clade 6 in this study, although the support for this<br />
grouping, and also the further clustering of clades 3 and 6<br />
with clade 7, was low. An alternative placement closer to the<br />
other predominantly papillate clades can therefore not be<br />
ruled out at present, although moderate support for a sistergroup<br />
relationship of clades 3 and 6 was also observed in<br />
the AU analysis. In line with Blair et al. (2008), P. quercina,<br />
which was considered a member of clade 3 in Cooke et al.<br />
(2000), was placed in clade 4, and is referred to as clade<br />
4.1 in this study, as this species was found to be basal to<br />
the group of the other members of clade 4 and the downy<br />
mildews. This placement received varying support in analysis<br />
of the full dataset and strong support in the reduced dataset.<br />
The predominantly non-papillate clades 6–10 were found<br />
predominantly in a basal position with respect to the crown<br />
group, providing evidence that the non-papillate stage<br />
might be ancestral, and the development of semi-papillate<br />
sporangia in clade 8b and clade 9 (sensu Blair et al. 2008)<br />
represents a homoplasy. Clade 9 (including clade 10) was<br />
found to be separated from the other Phytophthora clades<br />
with strong support and represented the most basal clade<br />
of Phytophthora. As was previously attested by Cooke et<br />
al. (2000), no obvious phylogenetic pattern with respect to<br />
temperature or climate adaptation can be observed from the<br />
phylogenetic analyses.<br />
Cooke et al. (2000) doubted if the species in these<br />
clades could be retained in Phytophthora and stated that it<br />
is likely that further investigation would lead to their exclusion<br />
from Phytophthora. As revealed in this study, paraphyly<br />
of Phytophthora is pronounced, rendering Phytophthora<br />
a typical example of a paraphyletic genus, with the most<br />
derived linages sharing some synapomorphies with downy<br />
mildews, while the more basal clades are more similar to<br />
Halophytophthora, Phytopythium and Pythium. This is similar<br />
to the situation in Peronosporales as a whole, for which Hulvey<br />
et al. (2010) recently proposed a broad circumscription of<br />
Peronosporaceae, encompassing all downy mildew genera,<br />
Halophytophthora, and Phytopythium, to avoid the description<br />
of several new, poorly differentiated families. If a similar option<br />
were chosen for the genus Phytophthora, this would mean an<br />
inclusion of all downy mildew genera and Phytophthora into a<br />
single genus. The oldest available name for this assemblage<br />
on genus level would be Peronospora (Corda 1837), which<br />
was described much earlier than Phytophthora (de Bary<br />
1876), thus, if Phytophthora were not conserved that would<br />
necessitate the inclusion of about 300 species of downy<br />
mildews, currently placed in other well-defined and widely<br />
accepted genera, e.g. Basidiophora, Bremia, Plasmopara,<br />
Peronosclerospora, Pseudoperonospora, and Scleropsora<br />
(Thines 2006, Voglmayr 2008), and about 100 species of<br />
Phytophthora (Waterhouse 1963, Erwin & Ribeiro 1996,<br />
Érsek & Ribeiro 2010) into this genus. This would not only be<br />
a nomenclatural nightmare but would also result in a highly<br />
heterogeneous group, encompassing species with divergent<br />
physiological, ecological, and morphological properties.<br />
For these reasons, but also because even more namechanges<br />
would be necessary, conservation of Phytophthora<br />
and an inclusion of all downy mildew genera (necessitating<br />
about 400–500 name changes for Peronospora alone), is<br />
not preferable. If this option were chosen, 700–800 names<br />
would have to be changed, including many well-known<br />
pathogens in the genera Bremia (e.g. Bremia lactucae),<br />
Plasmopara (e.g. Plasmopara viticola and Pl. halstedii),<br />
Hyaloperonospora (Hyaloperonospora brassicae, H.<br />
arabidopsidis, H. parasitica), and Peronospora (e.g. Pe.<br />
tabacina, Pe. destructor, Pe. effusa, Pe. farinosa, Pe. lamii).<br />
An alternative solution would be to resolve the paraphyly<br />
of this group by introducing new generic names where none<br />
existed for the lineages not belonging to the monophyletic<br />
subtree that includes Phytophthora infestans (the type<br />
species of Phytophthora). Judging from the results of this<br />
study, Phytophthora is at least six times paraphyletic as<br />
revealed by the phylogenetic investigations, but possibly<br />
seven times paraphyletic with respect to the downy mildews<br />
judging from the results obtained from the statistical tests.<br />
This would necessitate the introduction of new generic<br />
names (or the adoption of currently unused generic names)<br />
for clades 4.1, 4.2, 8, and the group (9, 10). In addition to<br />
these clusters, additional generic names would have to be<br />
introduced for groups formed by members of clades 2, 3,<br />
5, 6, and 7. In the phylogenetic analysis, while the groups<br />
(2, 5) and (3, 6, 7) were observed, their monophyly could<br />
not be ascertained; indeed, some support for alternative<br />
clusters (3, 6) and (5, 7), with clade 2 as an independent<br />
linage, was received in statistical tests. Several loci will<br />
need to be added in future phylogenetic studies to clarify<br />
the evolutionary relationships of these groups. Based on<br />
the current data, it can be assumed that Phytophthora is<br />
at least six, but possibly seven times paraphyletic with<br />
respect to downy mildews. Species of clade 1, which<br />
include the economically most important pathogen of the<br />
genus, Phytophthora infestans, as well as the well-known<br />
pathogens, P. nicotianae and P. cactorum, would retain their<br />
original names. This solution would need only a quarter<br />
of the name changes (less than 100) needed for the first<br />
option (inclusion of all downy mildew and Phytophthora<br />
species into Peronospora), and only about 15 % of the<br />
name changes that would be needed if Phytophthora were<br />
conserved and all downy mildews were transferred into<br />
this genus. In addition, it would leave the names of most<br />
of the most important pathogens of the Peronosporaceae<br />
unchanged, like Bremia lactucae, Hyaloperonospora<br />
brassicae, Phytophthora infestans, Plasmopara halstedii,<br />
Plasmopara viticola, Pseudoperonospora cubensis and<br />
ARTICLE<br />
volume 2 · no. 2<br />
169
Runge et al.<br />
ARTICLE<br />
Pseudoperonospora humuli. Therefore, we feel that this<br />
solution is to be preferred. But to introduce the new names<br />
for the clades outlined above will necessitate a search for<br />
characters defining synapomorphies for these groups, which<br />
might not be easy, judging from the apparent discrepancies<br />
between the morphological classification of Waterhouse<br />
(1963), and recent phylogenetic studies (Cooke et al. 2000,<br />
Kroon et al. 2004, Blair et al. 2008). Probably, these genera<br />
might have to be defined with the aid on DNA sequence<br />
synapomorphies, rather than only morphology. But retaining<br />
the usage of the generic name Phytophthora for all the at<br />
least six monophyletic groups between Halophytophthora<br />
and at the same time retaining the 19 downy mildew genera,<br />
would not only be contrary to the widely accepted idea of<br />
ideally having monophyletic taxa only, but also hamper the<br />
awareness of the unique evolution of these organisms,<br />
stepwise towards obligate biotrophy (Thines & Kamoun<br />
2010). For example, in terms of evolution, Phytophthora<br />
infestans is much closer to downy mildews than to P. sojae or<br />
even P. ramorum. But for the understanding of the evolution<br />
of obligate biotrophy, which is one of the most fascinating<br />
and fundamental evolutionary tipping points for any group of<br />
pathogens, it will be even more important to obtain genome<br />
sequences for members of the clades 4.1 and 4.2, which<br />
are apparently the closest relatives of the downy mildews,<br />
and of the neglected species of Phytophthora affecting<br />
Cyperaceae.<br />
Acknowledgements<br />
Fabian Runge is supported by a fellowship of the Ministry of Science<br />
and Education of Baden-Württemberg. This study was supported<br />
by the research funding programme “LOEWE – Landes-Offensive<br />
zur Entwicklung Wissenschaftlich-ökonomischer Exzellenz” of the<br />
Ministry of Higher Education, Research, and the Arts of Hesse. Work<br />
in the laboratory of BD was supported by a Michigan State University<br />
GREEEN grant (GR10-021)<br />
References<br />
Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller<br />
W, Lipman DJ (1997) Gapped BLAST and PSI-BLAST: A new<br />
generation of protein database search programs. Nucleic Acids<br />
Research 25: 3389–3402.<br />
Baxter L, Tripathy S, Ishaque N, Boot N, Cabral A, et al. (2010) Signatures<br />
of adaptation to obligate biotrophy in the Hyaloperonospora<br />
arabidopsidis genome. Science 330: 1149–1151.<br />
Belbahri L, Moralejo E, Calmin G, Oszako T, Garcia JA, et al. (2006)<br />
Phytophthora polonica, a new species isolated from declining<br />
Alnus glutinosa stands in Poland. FEMS Microbiology Letters<br />
261: 165–174.<br />
Blair JE, Coffey MD, Park SY, Geiser DM, Kang S (2008) A multilocus<br />
phylogeny for Phytophthora utilizing markers derived from<br />
complete genome sequences. Fungal Genetics and Biology 45:<br />
266–277.<br />
Brasier CM, Beales PA, Kirk SA, Denman S, Rose J (2005)<br />
Phytophthora kernoviae sp. nov., an invasive pathogen<br />
causing bleeding stem lesions on forest trees and foliar<br />
necrosis of ornamentals in the UK. Mycological Research 109:<br />
853–859.<br />
Cooke DEL, Drenth A, Duncan JM, Wagels G, Brasier CM (2000)<br />
A molecular phylogeny of Phytophthora and related oomycetes.<br />
Fungal Genetics and Biology 30: 17–32.<br />
Corda ACJ (1837) Icones Fungorum Hucusque Cognitorum. Vol. 1.<br />
Prague: J G Calve.<br />
De Bary A (1876) Researches into the nature of the potato fungus<br />
Phytophthora infestans. Journal of the Royal Agricultural Society<br />
of England, Series 2, 12: 239–269.<br />
Dick MW (2001) Straminipilous Fungi: systematics of the<br />
Peronosporomycetes including accounts of the marine<br />
straminipilous protists, the Plasmodiophorids and similar<br />
organisms. Dordrecht: Kluwer Academic.<br />
Dick MW, Wong PTW, Clark G (1984) The identity of the oomycete<br />
causing kikuyu yellows with a reclassification of the downy<br />
mildews. Botanical Journal of the Linnaean Society 89: 171–198.<br />
Dick MA, Dobbie K, Cooke DEL, Brasier CM (2006) Phytophthora<br />
captiosa sp. nov. and P. fallax sp. nov. causing crown dieback<br />
of Eucalyptus in New Zealand. Mycological Research 110: 393–<br />
404.<br />
Diéguez-Uribeondo J, García MA, Cerenius L, Kozubíková E,<br />
Ballesteros I, et al. (2009) Phylogenetic relationships among<br />
plant and animal parasites, and saprotrophs in Aphanomyces<br />
(Oomycetes). Fungal Genetics and Biology 46: 365–376.<br />
Érsek T, Ribeiro OK (2010) An annotated list of new Phytophthora<br />
species described post 1996. Acta Phytopathologica et<br />
Entomologica Hungarica 45: 251–266.<br />
Erwin DC, Ribeiro OK (1996) Phytophthora Diseases Worldwide. St<br />
Paul, MN: APS Press.<br />
Felsenstein J (1985) Confidence limits on phylogenies: an approach<br />
using the bootstrap. Evolution 39: 783–791.<br />
Gäumann EA (1952) The Fungi: a description of their morphological<br />
features and evolutionary development. New York: Hafner.<br />
Giresse X, Ahmed S, Richard-Cervera S, Delmotte F (2010)<br />
Development of new oomycete taxon-specific mitochondrial<br />
cytochrome b region primers for use in phylogenetic and<br />
phylogeographic studies. Journal of Phytopathology 158: 321–<br />
327.<br />
Göker M, Stamatakis A (2006) Maximum likelihood phylogenetic<br />
inference: an empirical comparison on a multi-locus dataset.<br />
German Conference on Bioinformatics 2006, Tübingen, Germany.<br />
Available online at http://citeseerx.ist.psu.edu/viewdoc/download<br />
?doi=10.1.1.89.201&rep=rep1&type=pdf.<br />
Göker M, Voglmayr H, Riethmüller A, Weiß M, Oberwinkler F (2003)<br />
Taxonomic aspects of Peronosporaceae inferred from Bayesian<br />
molecular phylogenetics. Canadian Journal of Botany 81: 672–<br />
683.<br />
Göker M, Voglmayr H, Riethmüller A, Oberwinkler F (2007) How do<br />
obligate parasites evolve? A multi-gene phylogenetic analysis of<br />
downy mildews. Fungal Genetics and Biology 44: 105–122.<br />
Haas BJ, Kamoun S, Zody MC, Jiang RH, Handsaker RE, et al. 2009.<br />
Genome sequence and comparative analysis of the Irish potato<br />
famine pathogen Phytophthora infestans. Nature 461: 393–398.<br />
170 ima fUNGUS
High degree of paraphyly in Phytophthora<br />
Hudspeth DSS, Stenger D, Hudspeth MES (2003) A cox2<br />
phylogenetic hypothesis for the downy mildew and white rusts.<br />
Fungal Diversity 13: 47–57.<br />
Huelsenbeck JP, Ronquist F (2001) MRBAYES: Bayesian inference<br />
of phylogenetic trees. Bioinformatics 17: 754–755.<br />
Hulvey J, Telle S, Nigrelli L, Lamour K, Thines M (2010)<br />
Salisapiliaceae – A new family of oomycetes from marsh grass<br />
litter of southeastern North America. Persoonia 25: 109–116.<br />
Katoh K, Kuma K-I, Toh H, Miyata T (2005) MAFFT version 5:<br />
Improvement in accuracy of multiple sequence alignment.<br />
Nucleic Acids Research 33: 511–518.<br />
Müller K (2004) PRAP—computation of Bremer support for large<br />
data sets. Molecular Phylogenetics and Evolution 31: 780–782.<br />
Payak MM, Renfro BL (1967) A new downy mildew disease of maize.<br />
Phytopathology 57: 394–397.<br />
Posada D, Crandall KA (1998) MODELTEST: Testing the model of<br />
DNA substitution. Bioinformatics 14: 817–818.<br />
Riethmüller A, Weiss M, Oberwinkler F (1999) Phylogenetic studies<br />
of Saprolegniomycetidae and related groups based on nuclear<br />
large subunit ribosomal DNA sequences. Canadian Journal of<br />
Botany 77: 1790–1800.<br />
Riethmüller A, Voglmayr H, Göker M, Weiss M, Oberwinkler F<br />
(2002) Phylogenetic relationships of the downy mildews<br />
(Peronosporales) and related groups based on nuclear large<br />
subunit ribosomal DNA sequences. Mycologia 94: 834–849.<br />
Shimodaira H (2002) An approximately unbiased test of phylogenetic<br />
tree selection. Systematic Biology 51: 492–508.<br />
Shimodaira H, Hasegawa M (2001) CONSEL: For assessing the<br />
confidence of phylogenetic tree selection. Bioinformatics 17:<br />
1246–1247.<br />
Stamatakis A, Hoover P, Rougemont J (2008) A Rapid Bootstrap<br />
Algorithm for the RAxML Web-Servers. Systematic Biology 75:<br />
758–771.<br />
Swofford DL (2002) PAUP*: phylogenetic analysis using parsimony<br />
(*and other methods), Version 4. Sunderland, MA: Sinauer<br />
Associates.<br />
Tamura K, Dudley J, Nei M, Kumar S (2007) MEGA4: Molecular<br />
Evolutionary Genetics Analysis (MEGA) software version 4.<br />
Molecular Biology and Evolution 24: 1596–1599.<br />
Thines M (2006) Evaluation of characters available from herbarium<br />
vouchers for the phylogeny of the downy mildew genera<br />
(Chromista, Peronosporales), with focus on scanning electron<br />
microscopy. Mycotaxon 97: 195–218.<br />
Thines M (2009) Bridging the gulf: Phytophthora and downy mildews<br />
are connected by rare grass parasites. PLoS ONE 4: e4790.<br />
Thines M, Kamoun S (2010) Oomycete-plant coevolution: Recent<br />
advances and future prospects. Current Opinion in Plant Biology<br />
13: 427–433.<br />
Thines M, Göker M, Oberwinkler F, Spring O (2007) A revision of<br />
Plasmopara penniseti, with implications for the host range of the<br />
downy mildews with pyriform haustoria (DMPH). Mycological<br />
Research 111: 1377–1385.<br />
Thines M, Göker M, Telle S, Ryley MJ, Mathur K, et al. (2008)<br />
Phylogenetic relationships of graminicolous downy mildews<br />
based on coxII sequence data. Mycological Research 112: 345–<br />
351.<br />
Thines M, Voglmayr H, Göker M (2009) Taxonomy and phylogeny of<br />
the downy mildews (Peronosporaceae). In: Oomycete Genetics<br />
and Genomics: biology, interactions with plants and animals, and<br />
toolbox (K Lamour & S Kamoun, eds): 47–75. Hoboken, NJ: J<br />
Wiley.<br />
Tian M, Win J, Savory E, Burkhardt A, Held M, Brandizzi F, Day B<br />
(2011) 454 genome sequencing of Pseudoperonospora cubensis<br />
reveals effector proteins with a QXLR translocation motif.<br />
Molecular Plant-Microbe Interactions 24: 543–553.<br />
Tiwari MM, Arya HC (1969) Sclerospora graminicola - axenic culture.<br />
Science 163: 291–292.<br />
Tokura R (1975) Axenic or artificial culture of the downy mildew fungi<br />
of gramineous plants. Tropical Agriculture Research Series 8:<br />
57–60.<br />
Tyler BM, Tripathy S, Zhang X, Dehal P, Jiang RH, et al. (2006)<br />
Phytophthora genome sequences uncover evolutionary origins<br />
and mechanisms of pathogenesis. Science 313: 1261–1266.<br />
Voglmayr H (2008) Progress and challenges in systematics of downy<br />
mildews and white blister rusts: new insights from genes and<br />
morphology. European Journal of Plant Pathology 122: 3–18.<br />
Waterhouse GM (1963) Key to the species of Phytophthora de Bary.<br />
Mycological Papers 92: 1–22.<br />
Waterhouse GM (1973) Peronosporales. In: The Fungi: an advanced<br />
treatise. Vol. 4B. A Taxonomic Review with Keys: basidiomycetes<br />
and lower fungi (GC Ainsworth, FK Sparrow, AS Sussman, eds):<br />
165–183. New York: Academic Press.<br />
ARTICLE<br />
volume 2 · no. 2<br />
171
ARTICLE<br />
<br />
ima fUNGUS
doi:10.5598/imafungus.2011.02.02.08 <strong>IMA</strong> <strong>Fungus</strong> · volume 2 · no 2: 173–175<br />
Validation and justification of the phylum name Cryptomycota phyl. nov.<br />
Meredith D.M. Jones 1,2 , Thomas A. Richards 1,2 , David L. Hawksworth 3 , and David Bass 2<br />
1<br />
School of Biosciences, University of Exeter, Exeter EX4 4QD, UK; corresponding author e-mail: d.bass@nhm.ac.uk<br />
2<br />
Department of Zoology, Natural History Museum, Cromwell Road, London SW7 5BD, UK<br />
3<br />
Departamento de Biología Vegetal II, Facultad de Farmacia, Universidad Complutense de Madrid, Plaza Ramón y Cajal, 28040 Madrid,<br />
Spain; and Department of Botany, Natural History Museum, Cromwell Road, London SW7 5BD, UK<br />
ARTICLE<br />
Abstract: The recently proposed new phylum name Cryptomycota phyl. nov. is validly published in order to facilitate its<br />
use in future discussions of the ecology, biology, and phylogenetic relationships of the constituent organisms. This name<br />
is preferred over the previously tentatively proposed “Rozellida” as new data suggest that the life-style and morphology<br />
of Rozella is not representative of the large radiation to which it and other Cryptomycota belong. Furthermore, taxa at<br />
higher ranks such as phylum are considered better not based on individual names of included genera, but rather on some<br />
special characteristics – in this case the cryptic nature of this group and that they were initially revealed by molecular<br />
methods rather than morphological discovery. If the group were later viewed as a member of a different kingdom, the<br />
name should be retained to indicate its fungal affinities, as is the practice for other fungal-like protist groups.<br />
Key words:<br />
chitin<br />
chytrid<br />
Fungi<br />
phylogeny<br />
Rozella<br />
Rozellida<br />
Article info: Submitted 26 September 2011; Accepted 2 November 2011; Published 11 November 2011.<br />
Introduction<br />
The designation “cryptomycota” was introduced by Jones<br />
et al. (2011) to accommodate a well-supported clade (using<br />
ribosomal DNA (rDNA) phylogenies) of organisms putatively<br />
branching deep within the fungal radiation. The rank of phylum<br />
is the most appropriate for this group as current results show<br />
that it has fungal characteristics but is distinct from other fungi<br />
in not having a chitin-rich cell wall in the major stages of its lifecycle<br />
so far identified, including putative trophic interactions.<br />
However, Cryptomycota was not validly published as a<br />
scientific name in that work as no Latin diagnosis was provided<br />
(McNeill et al. 2006: Art. 36). A Latin diagnosis is provided here<br />
in order to formally establish the name. In addition, comments<br />
are made on our decision to introduce this name rather than<br />
take up the earlier informal name “Rozellida”, and on the<br />
distinctive features of the phylum and its position.<br />
TAXONOMY<br />
Cryptomycota M. D. M. Jones & T. A. Richards, phyl.<br />
nov.<br />
MycoBank MB563383<br />
Etymology: crypto- – hidden; and -mycota, a phylum of fungi.<br />
Fungi unicellulares, zoosporis unicellularis, uniflagellatibus, flagellis<br />
microtubularis, cystes sine tunica chitinosa vel cellulosa. Consortia<br />
epibiontica formata.<br />
Fungi unicellular, zoospores single-celled with a single<br />
microtubular flagellum, and cysts without a chitin/cellulose<br />
cell wall. Forming epibiontic associations.<br />
Representatives: GenBank accession nos AJ130857,<br />
AJ130849.1, AJ130850, FJ687265, FJ687267 and FJ687268,<br />
and Rozella.<br />
Illustrations: Jones et al. (2011: figs 1d, 2a–e).<br />
DISCUSSION<br />
It has been demonstrated that Rozella occupies a deep<br />
branching position in phylogenetic analyses of kingdom<br />
Fungi (James et al. 2006a, b), although bootstrap support for<br />
this relationship is inconsistent and often weak in the most<br />
comprehensively sampled phylogenies (James et al. 2006a,<br />
b, Jones et al. 2011). The name “Rozellida” was coined by<br />
Lara et al. (2010) to accommodate Rozella and a number of<br />
environmental sequences that form a distinct clade, but we<br />
refer to this group henceforth as Cryptomycota for reasons<br />
indicated below. Jones et al. (2011) showed that Cryptomycota<br />
are more diverse than previously recognised and that the<br />
molecular diversity of this group may be as diverse as the rest<br />
of the known Fungi according to rDNA gene markers.<br />
Members of Cryptomycota are found in freshwater, soil,<br />
sediment, and some marine habitats. Jones et al. (2011) used<br />
© 2011 International Mycological Association<br />
You are free to share - to copy, distribute and transmit the work, under the following conditions:<br />
Attribution:<br />
You must attribute the work in the manner specified by the author or licensor (but not in any way that suggests that they endorse you or your use of the work).<br />
Non-commercial: You may not use this work for commercial purposes.<br />
No derivative works: You may not alter, transform, or build upon this work.<br />
For any reuse or distribution, you must make clear to others the license terms of this work, which can be found at http://creativecommons.org/licenses/by-nc-nd/3.0/legalcode. Any of the above conditions can be waived if you get<br />
permission from the copyright holder. Nothing in this license impairs or restricts the author’s moral rights.<br />
volume 2 · no. 2 173
Jones et al.<br />
ARTICLE<br />
lineage-specific fluorescence in situ hybridization (FISH), cell<br />
wall stains, and immuno-fluorescence staining to show two<br />
distinct lineages within Cryptomycota, which comprised ovoid<br />
cells of ca. 5 µm diam, existing in at least three morphologies<br />
in freshwater environments: uniflagellate zoospores, more<br />
variably-shaped cells without flagella attached to other eukaryotic<br />
microscopic organisms (e.g. diatom hosts), and non-flagellate<br />
cysts. None of these stages were shown to possess a chitin or<br />
cellulose wall, although other life-cycle phases with a chitin and/<br />
or cellulose cell wall may remain undetected. A chitin cell wall is<br />
sometimes cited as defining feature of kingdom Fungi, although<br />
we note that this is not a reliable diagnostic feature as distantly<br />
related protist groups also possess chitin on their cell surface<br />
(e.g. Kneipp et al. 1998).<br />
The name “Rozellida” was applied to this phylum by Lara<br />
et al. (2010) but in an informal way between inverted commas<br />
and with no formal diagnosis. The ICZN (International Code<br />
of Zoological Nomenclature; International Commission on<br />
Zoological Nomenclature 1999) does not apply to names<br />
above the rank of family-group, but if it were in those ranks<br />
it would be viewed as unavailable as a conditional name<br />
(Art. 15.1). For names introduced under the ICZN which<br />
later are found to belong to Fungi, the ICN (International<br />
Code of Nomenclature for algae, fungi, and plants) now<br />
accepts them as available under Art. 45.4 (as revised at the<br />
Melbourne Congress in July 2011; McNeill et al. 2011). Thus,<br />
no Latin diagnosis was required, as it was for fungal names<br />
introduced between 1935 and 1 January 2012). However, we<br />
are inclined not to accept “Rozellida” because of the use of<br />
the inverted commas suggesting the usage was a tentative<br />
suggestion and in any case note that it is not mandatory to<br />
follow the principle of priority of publication for names above<br />
the rank of family (ICN) or family group (ICZN). Indeed, the<br />
ICZN does not cover ranks higher than family group.<br />
We decided that it would be better not to definitely<br />
establish a name based on Rozella for several reasons:<br />
(1) The fungal termination to be used for names in the<br />
rank of phylum is “-mycota” under the ICN (McNeill et al. 2006;<br />
Art 16.4), and that termination has also been used for phyla<br />
traditionally studied by mycologists but which are no longer<br />
considered Fungi but placed in other kingdoms. Examples<br />
include Hyphochytriomycota R.H. Whittaker (Whittaker 1969:<br />
154) now placed in Straminipila M.W. Dick 2001, Myxomycota<br />
Bold (Bold 1957: 152) for slime moulds in the Protozoa, and<br />
Oomycota Arx (Arx 1967: 16) for fungal analogues in the<br />
Straminipila. This practice has been employed in standard<br />
reference works (e.g. Kirk et al. 2001) and also the most<br />
recent textbooks (e.g. Moore et al. 2011).<br />
(2) Cryptomycota represent a very diverse radiation,<br />
potentially equivalent to or larger than the rest of the known<br />
fungi. Of the three lineages within the radiation for which<br />
morphological data exist, Rozella appears to be exceptional<br />
in that it is primarily an intracellular parasite; indeed the<br />
possession of intracellular sporangia is included in the<br />
generic description of Rozella species (Held 1981). To extend<br />
the implication of this life-cycle characteristic across the rest<br />
of the radiation – where there is no evidence of this lifecycle<br />
characteristic – would be misleading. Lara et al. (2010)<br />
were also hesitant commending the use of the proposed<br />
name “between quotation marks until morphological and/or<br />
ultrastructural synapomorphies are defined to diagnose and<br />
validate this entire group”. Jones et al. (2011) demonstrate<br />
that this key characteristic of Rozella does not seem to extend<br />
across the whole group and therefore the name “Rozellida” is<br />
not representative of the group as a whole.<br />
(3) It is important to recognize that our current knowledge<br />
of the life stages of the newly discovered Cryptomycota<br />
and of Rozella is very incomplete. As Jones et al. (2011)<br />
suggest, chitin may be present in the walls of some currently<br />
unknown Cryptomycota life-cycle stage(s) and/or present in<br />
uncharacterized lineages within Cryptomycota, and even in<br />
currently unknown stages in Rozella. It would be premature,<br />
therefore, to separate Cryptomycota from the kingdom Fungi<br />
on the single character that they do not possess chitin walls<br />
(which, as mentioned above is not diagnostic for Fungi).<br />
(4) Cryptomycota have some strong resemblances to<br />
Chytridiomycota (‘chytrids’) in both structure (e.g. flagellar<br />
apparatus) and ecology, if not in cell wall chemistry. There is<br />
no agreed defining non-molecular characteristic for identifying<br />
the boundaries of kingdom Fungi. Therefore, as several other<br />
key characteristics are shared by Cryptomycota and some<br />
Fungi, the former are most sensibly and parsimoniously<br />
considered as belonging to the latter as they form the closest<br />
branches on phylogenetic trees (James et al. 2006a, b,<br />
Lara et al. 2010, Jones et al. 2011). This stance is entirely<br />
consistent with the historical position regarding Rozella: for<br />
the last 40 years leading mycologists have classified this<br />
genus within Fungi (e.g. Held, 1981; Kirk et al. 2008).<br />
(5) Cryptomycota (including Rozella) consistently branch<br />
with Fungi in all phylogenies so far constructed. However, their<br />
position as the primary branch within fungi is much weaker<br />
(e.g. James et al. 2006; Jones et al. 2011). Indeed, they could<br />
actually occupy a higher branching position within Fungi. If<br />
this is the case, their lack of some traditionally diagnostic<br />
fungal features such as a chitin cell wall may be the result of<br />
secondary losses, which would not preclude them from being<br />
considered Fungi. In this case, excluding Cryptomycota from<br />
the Fungi could potentially make the rest of fungi paraphyletic<br />
– a highly undesirable and not logically sustainable situation.<br />
In the absence of a strong morphological argument to exclude<br />
this group from the fungal kingdom – we must therefore look<br />
to the only available data, which is phylogenetic, and argues<br />
that Cryptomycota are most reasonably considered to be<br />
within Fungi.<br />
(6) Consequently, we agree with Lara et al. (2010) that<br />
there are sound reasons for considering Rozella (and now<br />
we suggest other Cryptomycota) as Fungi. Whether or not<br />
Cryptomycota other than Rozella prove to be phagocytotic<br />
(which in itself would not be a sufficiently strongly deterministic<br />
trait for inclusion in – or exclusion from – Fungi, as some<br />
plant lineages and oomycetes have also lost phagotrophy),<br />
their chytrid-like uniflagellate zoospore stage and particularly<br />
their phylogenetic position argue most parsimoniously for a<br />
fungal affiliation.<br />
174 ima fUNGUS
Validation of the phylum name Cryptomycota<br />
(7) The names used for taxa at the highest ranks, such as<br />
phylum, are better not based on names of included genera,<br />
but rather on some special characteristic, as is the case with,<br />
for example, the phyla Ascomycota and Basidiomycota. In<br />
this way the names immediately convey some feature of<br />
the taxon. In this case, we highlight the cryptic nature of<br />
Cryptomycota in that they were hidden from science until<br />
revealed by molecular methods rather than morphological<br />
discovery.<br />
In conclusion, we consider the formal validation of the<br />
name Cryptomycota to be justified, and commend it for use<br />
for this group of organisms as it emphasises the fungal affinity<br />
and attributes of the organisms so far known within this<br />
group. Even if in some future classification these organisms<br />
were placed outside the Fungi, we consider the name should<br />
be retained to reflect their nature as fungal analogues.<br />
ACKNOWLEDGEMENTS<br />
We thank Ramon Massana, Irene Forn, Caterina Gadelha, and Martin<br />
Egan for useful discussions, and Joost A Stalpers for checking the<br />
Latin diagnosis. D. L. H. acknowledges support from the Ministerio<br />
de Ciencia e Innovación in Spain (project CGL 2008-01600).<br />
REFERENCES<br />
Arx JA von (1967) Pilzkunde: Ein kurzer Abriss der Mykologie under<br />
besonderer Berücksichtigung der Pilze in Reinkultu. Lehre: J<br />
Cramer.<br />
Bold HA (1957) Morphology of Plants. New York: Harper Row.<br />
Held AA (1981) Rozella and Rozellopsis: naked endoparasitic fungi<br />
which dress up as their hosts. Botanical Review 47: 451–515.<br />
International Commission on Zoological Nomenclature (1999)<br />
International Code of Zoological Nomenclature. 4 th edn. London:<br />
International Trust for Zoological Nomenclature.<br />
James TY, Kauff F, Schoch CL, Matheny PB, Hofstetter V, Cox CJ<br />
[and 64 others], (2006a) Reconstructing the early evolution of<br />
Fungi using a six-gene phylogeny. Nature 443: 818–822.<br />
James TY, Letcher PM, Longcore JE, Mozley-Standridge SE, Porter<br />
D, Powell MJ, Griffith GW, Vilgalys R (2006b) A molecular<br />
phylogeny of the flagellated fungi (Chytridiomycota) and<br />
description of a new phylum (Blastocladiomycota). Mycologia<br />
98: 860–871.<br />
Jones MDM, Forn I, Gadelha C, Egan MJ, Bass D, Massana R,<br />
Richards TA (2011) Discovery of novel intermediate forms<br />
redefines the fungal tree of life. Nature 474: 200–203.<br />
Kirk PM, Cannon PF, David JC, Stalpers JA (eds) (2001) Ainsworth<br />
& Bisby’s Dictionary of the Fungi. 9 th edn. Wallingford: CABI<br />
Publishing.<br />
Kirk PM, Cannon PF, Minter DW, Stalpers JA (eds) (2008) Ainsworth<br />
& Bisby’s Dictionary of the Fungi. 10 th edn. Wallingford: CABI<br />
Publishing.<br />
Kneipp LF, Andrade AF, de Souza W, Angluster J, Alviano CS,<br />
Travassos LR (1998) Trichomonas vaginalis and Trichomonas<br />
foetus: expression of chitin at the cell surface. Experimental<br />
Parasitology 89: 195–204.<br />
Lara E, Moreira D, López-Garcia P (2010) The environmental clade<br />
LKM11 and Rozella form the deepest branching clade of Fungi.<br />
Protist 161: 116–121.<br />
McNeill J, Turland NJ, Monro A, Lepschi B (2011) XVIII International<br />
Botanical Congress: preliminary mail vote and report of congress<br />
action on nomenclature proposals. Taxon 60: 1507–1520.<br />
Moore D, Robson GD, Trinci APJ (2011) 21 st Century Guidebook to<br />
Fungi. Cambridge: Cambridge University Press.<br />
Whittaker RH (1969) New concepts of kingdoms of organisms.<br />
Science 163: 150–160.<br />
ARTICLE<br />
volume 2 · no. 2<br />
175
ARTICLE<br />
<br />
ima fUNGUS
doi:10.5598/imafungus.2011.02.02.09 <strong>IMA</strong> <strong>Fungus</strong> · volume 2 · no 2: 177–189<br />
Molecular techniques for pathogen identification and fungus detection in the<br />
environment<br />
Clement K.M. Tsui 1 , James Woodhall 2 , Wen Chen 3 , C. André Lévesque 3 , Anna Lau 4 *, Cor D. Schoen 5 , Christiane Baschien 6** ,<br />
Mohammad J. Najafzadeh 7,8 , and G. Sybren de Hoog 7<br />
ARTICLE<br />
1<br />
Department of Forest Sciences, The University of British Columbia, Vancouver, BC, V6T 1Z4, Canada; corresponding author e-mail:<br />
clementsui@gmail.com<br />
2<br />
The Food and Environment Research Agency, Sand Hutton, York YO41 1LZ, UK<br />
3<br />
Central Experimental Farm, Agriculture and Agri-Food Canada, Ottawa, Canada, K1A OC6<br />
4<br />
Centre for Infectious Diseases and Microbiology and the University of Sydney, Westmead Hospital, Westmead, NSW 2145, Australia<br />
5<br />
Plant Research International, Business Unit Bio-Interactions and Plant Health, PO Box 16, 6700 AA, Wageningen, The Netherlands<br />
6<br />
Technische Universität Berlin, Environmental Microbiology, Sekr. FR1-2, Franklinstrasse 29, 10587 Berlin, Germany<br />
7<br />
CBS-KNAW Fungal Biodiversity Centre, Uppsalalaan 8, 3584 CT Utrecht, The Netherlands<br />
8<br />
Department of Parasitology and Mycology, School of Medicine, Mashhad University of Medical Sciences, Mashhad, Iran<br />
*Current mailing address: Department of Laboratory Medicine, 10 Center Drive, National Institutes of Health, Bethesda, MD 20892, USA<br />
**Current mailing address: Federal Environment Agency Germany, Corrensplatz 1, 14195 Berlin, Germany<br />
Abstract: Many species of fungi can cause disease in plants, animals and humans. Accurate and robust<br />
detection and quantification of fungi is essential for diagnosis, modeling and surveillance. Also direct<br />
detection of fungi enables a deeper understanding of natural microbial communities, particularly as a great<br />
many fungi are difficult or impossible to cultivate. In the last decade, effective amplification platforms, probe<br />
development and various quantitative PCR technologies have revolutionized research on fungal detection<br />
and identification. Examples of the latest technology in fungal detection and differentiation are discussed<br />
here.<br />
Key words:<br />
FISH<br />
LAMP<br />
macroarray<br />
medical mycology<br />
molecular diagnostics<br />
molecular ecology<br />
padlock probe<br />
pathogenic fungi<br />
plant pathology<br />
rolling circle amplification<br />
Article info: Submitted 17 October 2011; Accepted 3 November 2011; Published 18 November 2011.<br />
Introduction<br />
Fungi represent the greatest eukaryotic diversity on earth and<br />
they are among the primary decomposers in ecosystems. It is<br />
conservatively estimated that 1.5 million species of fungi exist<br />
(Hawksworth 1991). Many fungal species are important plant<br />
and human pathogens (Agrios 2005). Rapid and accurate<br />
detection of fungal pathogens to species or strain level is<br />
often essential for disease surveillance and implementing a<br />
disease management strategy. Developing direct detection<br />
assays is challenging because fungal pathogens can exist<br />
as multiple species complexes or at very low concentration<br />
in clinical and natural environments. Different molecular<br />
genotypes/varieties can also exist within species, and may<br />
have different pathogenic profiles and virulence levels to<br />
the host. In addition, unculturable and non-sporulating<br />
fungi remain a major challenge when studying biotrophic,<br />
endophytic, and mycorrhizal groups. Therefore, novel<br />
techniques are required when attempting to detect fungi in<br />
the environment.<br />
Increasingly molecular techniques are employed in<br />
studies requiring the detection of fungi in the environment.<br />
In this paper, based on presentations at a Special<br />
Interest Group meeting convened during the International<br />
Mycological Congress (IMC9) in Edinburgh, UK, August<br />
2010, some of the latest diagnostic techniques employed<br />
in the detection of fungi, including fluorescence in situ<br />
hybridisation (FISH), DNA array technology, Multiplex<br />
tandem PCR, and Padlock probe technology with<br />
rolling circle amplification and loop-mediated isothermal<br />
amplification (LAMP) were discussed. The importance of<br />
DNA extraction and sampling methodologies were also<br />
briefly discussed.<br />
© 2011 International Mycological Association<br />
You are free to share - to copy, distribute and transmit the work, under the following conditions:<br />
Attribution:<br />
You must attribute the work in the manner specified by the author or licensor (but not in any way that suggests that they endorse you or your use of the work).<br />
Non-commercial: You may not use this work for commercial purposes.<br />
No derivative works: You may not alter, transform, or build upon this work.<br />
For any reuse or distribution, you must make clear to others the license terms of this work, which can be found at http://creativecommons.org/licenses/by-nc-nd/3.0/legalcode. Any of the above conditions can be waived if you get<br />
permission from the copyright holder. Nothing in this license impairs or restricts the author’s moral rights.<br />
volume 2 · no. 2 177
Tsui et al.<br />
ARTICLE<br />
GENERAL CONSIDERATIONS<br />
Suitable methodologies to isolate nucleic acids from the<br />
enormous range of habitats that fungi can occupy are crucial<br />
to all molecular detection methods. Critical to the successful<br />
isolation of nucleic acids are the methods used for extraction<br />
and sample preparation, as well as the sampling strategy<br />
employed.<br />
DNA extraction methods<br />
An enormous variety of nucleic acid extraction methods<br />
is available. For many applications, commercial kits are<br />
available, but these are not always suitable. For example, in<br />
the case of soil, none of the kits presently available are able<br />
to extract efficiently using sample sizes typically required<br />
(see below). Therefore, with unusual and difficult sample<br />
types, customized methods are typically employed. Key to<br />
this is the cell disruption/homogenisation step in the presence<br />
of an appropriate buffer (typically a CTAB or guanidinium<br />
thiocyanate based buffer). A wide variety of methods can be<br />
used, including specialist equipments such as a planetary<br />
ball mill (Brierley et al. 2009), appropriate bead beaters,<br />
pressure cycling technology (Okubara et al. 2007), and novel<br />
approaches using equipment such as a paint shaker for soil<br />
samples (Reeleder et al. 2003), or even a conventional food<br />
blender for plant and food samples. All cell disruption steps are<br />
potentially damaging to the nucleic acid quantity and quality,<br />
either through direct shearing of the nucleic acid, or through<br />
the co-extraction of humic acids (an inhibitor of PCR) present<br />
in plant and soil samples. Therefore, methods which provide an<br />
adequate level of cell disruption to enable satisfactory nucleic<br />
acid extraction, but without too much nucleic acid damage or<br />
release of potential PCR inhibitors, are required.<br />
Following cell disruption and subsequent extraction steps,<br />
the nucleic acid can be purified using standard methods,<br />
including pelleting, silica membrane spin filter, and silicamagnetic<br />
particle separation. When selecting a purification<br />
step, consideration needs to be given to the nucleic acid<br />
quality/quantity required, cost, speed of extraction and ability<br />
to automate any process. The method of analysing the nucleic<br />
acid will determine what purification method is used. For<br />
example, DNA profiling and metagenomic studies using next<br />
generation sequencing approaches will require pure, high<br />
quality nucleic acid extracts. Conversely, cruder, less pure<br />
extracts may be used where real-time PCR is used for routine<br />
diagnostic purposes where the emphasis is on nucleic acid<br />
quantity, speed and cost of extraction because the downstream<br />
application is more tolerant of impurities in the DNA extract.<br />
Sample preparation<br />
Innovative sample preparation can enhance chances of<br />
detection. For example, organic matter can be removed<br />
from soil and processed separately for fungal targets which<br />
survive solely in that component of the soil. Sclerotiumforming<br />
pathogens may be separated from the rest of the soil<br />
by sieving or floating prior to nucleic acid extraction. These<br />
approaches effectively concentrate the fungal target, thereby<br />
increasing the chance of successful detection. Where<br />
appropriate, a baiting approach could be used in conjunction<br />
with nucleic acid extraction and PCR. This has the added<br />
benefit of confirming the target is viable.<br />
Sampling strategy<br />
In addition, an appropriate and statistically robust sampling<br />
strategy should be utilised. This will vary between fungal<br />
targets, depending upon the biology (and possibly<br />
epidemiology) of the organism to be detected. For example, a<br />
‘W’ sampling strategy across an area may not be appropriate<br />
for a target with a highly clustered distribution, and a grid<br />
sampling approach should be used instead. The individual<br />
sample size processed for DNA extraction should also be<br />
large enough to be biologically relevant (Ophel-Keller et al.<br />
2008). A wide variety of methods exist, and no one approach<br />
is likely to be suitable for all targets. Knowledge of the biology<br />
of the target species is essential for designing and determining<br />
the optimum sampling and extraction methodology in any<br />
particular case.<br />
Sequencing independent methods<br />
DNA sequencing of the internal transcribed spacer (ITS) and<br />
large subunit (LSU) regions of rRNA, followed by comparative<br />
sequence analysis, has been the ‘gold standard’ for molecular<br />
identification of most fungi, particularly of culturable fungi. This<br />
strategy is fast and accurate, but is dependent on sequence<br />
quality in existing reference databases. In many recently<br />
evolved fungal groups, however, the ribosomal genes are<br />
insufficiently variable, and sequencing of additional genes<br />
is necessary. Costs could also be a challenge for routine<br />
diagnostic laboratories, and therefore various sequencing<br />
independent methods that are available can be used.<br />
Fluorescent in situ hybridisation<br />
Since fungi are ubiquitous, it is important to understand their<br />
biodiversity and abundance, as well as their ecological roles<br />
in different habitats, such as soil, decaying leaves and wood,<br />
and also indoor environments or humans. Indeed, it is critical<br />
to identify the metabolically active species (‘real players’)<br />
in communities or ecosystems in order to understand<br />
the ecosystem processes that involve fungi. PCR based<br />
methods, such as fingerprinting or molecular cloning, do not<br />
discriminate between ‘real players’ and fungi that are dormant.<br />
Also, DNA and RNA extraction protocols may be biased due<br />
to rigid cell walls of fungal hyphae. Immuno-staining provides<br />
a means to identify species in situ but is extremely laborious<br />
in the experimental preparation. All these kinds of technical<br />
difficulties, can, however, be overcome by fluorescence in<br />
situ hybridisation (FISH).<br />
FISH is a powerful method for the in situ detection of<br />
active growing organisms in environmental samples (Amann<br />
et al. 1995). The technique can visualize the precise location<br />
178<br />
<br />
ima fUNGUS
Molecular identification and detection of fungi<br />
ARTICLE<br />
Fig. 1. Examples of fluorescence in situ hybridisation. A. Clavariopsis aquatica growing on aquatic leaf litter, probed with 18 rRNA-targeted<br />
MY1574 domain specific probe (from Baschien et al. 2008). B. Accessibility of Tetracladium marchalianum conidia for FISH 28SrRNA-targeted<br />
species specific probe TmarchB10 (modified from Baschien et al. 2008).<br />
of particular DNA or RNA sequences in the cytoplasm,<br />
organelles, or nuclei of biological materials. As a result, the<br />
technique can detect metabolically active fungi directly in the<br />
environment without cultivation when RNA is present. The<br />
spatial distribution of growing mycelia on or within colonized<br />
substrata can also be investigated (Li et al. 1996, Baschien et<br />
al. 2001, McArthur et al. 2001, Robin et al. 1986).<br />
The major step of FISH involves the preparation of<br />
biological materials or environmental samples, and the<br />
labelling (incorporation of a fluorescent label/marker) of a<br />
nucleic acid sequence to form a probe. Then, under controlled<br />
experimental conditions, the probe is hybridized to the DNA<br />
or RNA in biological materials to form a double-stranded<br />
molecule. Finally, the sites of hybridization are detected<br />
and visualized. FISH is commonly used in the ‘top-bottom<br />
approach’ research followed with feedback to the top level<br />
(Amann et al. 1995). The molecular biology and genetics<br />
of targeted organism is well-understood, and the probe is<br />
designed based on the nucleotide sequence which has been<br />
well characterized.<br />
FISH probes often target sequences of ribosomal RNA<br />
or mitochondrial genes because they are abundant in<br />
sequence databases and in multiple copies in each cell. The<br />
probes can be designed by computer-assisted search from<br />
target organisms for “signatures”. These signature regions<br />
can be specific at different phylogenetic levels depending<br />
on the variability of the target molecule sequences. The<br />
probe comprises a short sequence, ranging from 15 to<br />
20 nucleotides, that is specific for one or several taxa at<br />
species, genus, or higher taxonomic ranks. The probe or<br />
oligonucleotide is then labelled with a fluorochrome, for<br />
example a carboindocyanine dye (CY3).<br />
A particular attraction is that FISH probes can be applied<br />
to formaldehyde or ethanol fixed environmental samples, or<br />
to cultures. At the ribosomes, the probe will specifically anneal<br />
to its complementary sequence resulting in a heteroduplex.<br />
After incubation, a washing step is crucial to discriminate the<br />
target from non-target organisms. Probes that do not bind<br />
to an rRNA sequence will be washed away, resulting in a<br />
fluorescent signal of exclusively target organisms. The probe<br />
conferred signal is correlated with the ribosome content, and<br />
therefore increases in cells with higher metabolic activity.<br />
The first FISH probe targeting a living fungus was<br />
designed by Li et al. (1996) for Aureobasidium pullulans<br />
on the phylloplane of apple seedlings; this was the first<br />
time that a living fungus had been visualized by FISH.<br />
Examples of situations in which the method has been used<br />
are very varied. Fungi belonging to Eurotiomycetes were<br />
demonstrated to be more abundant than Dothideomycetes in<br />
an extremely acidic mine drainage using 18S rRNA-targeted<br />
probes (Baker et al. 2004). Baschien et al. (2008) designed<br />
nine taxon-specific probes targeting the 18S or 28S rRNA<br />
for the detection of aquatic hyphomycetes in leaf litter (Fig.<br />
1A, B). Domain specific probes were also used to detect<br />
active fungi in mice (Scupham et al. 2006) and in waste water<br />
sewage granule biofilms (Weber et al. 2007). Most probes<br />
have been designed to target the 18S or 28S rRNA gene, and<br />
their specificity needs to be kept under review as sequence<br />
databases expand.<br />
Several factors can influence the efficiency of FISH, for<br />
example the sterical and electrostatical properties of rRNA,<br />
the features of the probe, and hybridisation conditions such<br />
as the fixation method, buffers, the stringency of probe<br />
binding, and incubation time. Inacio et al. (2003) investigated<br />
volume 2 · no. 2<br />
179
Tsui et al.<br />
ARTICLE<br />
the different accessibility of the rRNA molecule for FISH<br />
probes. They designed 32 different probes that targeted the<br />
26S (large subunit) of rRNA for yeasts, and investigated the<br />
conferred signal intensity generated from the probes. They<br />
concluded that the prerequisite for successful FISH is the<br />
specificity of probes (Inácio et al. 2003).<br />
Limitations of the FISH method can include fungal and<br />
substrate inherent autofluorescence, insufficient permeability<br />
of cell walls, non-specific binding of probes, and low ribosome<br />
contents. Autofluorescence of fungi can also lead to false<br />
positive fluorescence signals. A fluorescence scan conducted<br />
by confocal laser scanning microscopy revealed that many<br />
freshwater fungi emit a green autofluorescence (Baschien et<br />
al. 2001), and, consequently, a green-labelled FISH probe<br />
could not be used. It is, therefore, important to check for<br />
autofluorescence of both target and non-target organisms<br />
before selecting the fluorochrome for probe labelling.<br />
Environmental samples and substrates such as leaves, wood,<br />
and animal t<strong>issue</strong>s, also emit strong autofluorescence in<br />
several wavelengths at the same time, interfering with probe<br />
signals. However, the pin-hole technique of a laser scanning<br />
microscope is helpful in reducing background emissions<br />
compared to conventional epifluorescence (Baschien et al.<br />
2001).<br />
Low or no permeability of fungal cell walls can lead to<br />
weak or no signals because of the failure of FISH probes to<br />
penetrate rigid cell walls (Brul et al. 1997). One possible way<br />
to overcome this problem is to use cell wall lysing enzymes,<br />
such as chitinases and glucanases. A more elegant method<br />
is the use of peptide nucleic acid (PNA) probes. PNA probes<br />
are mimics in which the negatively charged sugar-backbone<br />
of DNA is replaced with a non-charged polyamide backbone.<br />
PNA probes share the same nucleotide bases as the<br />
conventional DNA probes. However, PNA probes penetrate<br />
cell walls more effectively due to their neutrality and they do<br />
not have to overcome the destabilizing electrostatic repulsion<br />
during hybridisation (Wilson et al. 2005, Shepard et al. 2008).<br />
Low ribosome contents can arise from either a scarcity of<br />
potential substrate in oligotrophic habitats, or the presence of<br />
components inhibiting fungal growth. However, the Catalysed<br />
Reporter Deposition FISH (CARD-FISH) is a variant of the<br />
FISH method designed to enhance probe conferred signals.<br />
The major difference is that the probes are labelled with horseradish-oxidase<br />
instead of a fluorochrome. Fluorescencelabelled<br />
tyramide is then added to the cells after hybridisation.<br />
The horse-radish-oxidase catalyses the deposition of the<br />
reporter, and this reaction leads to 20 to 30 fold stronger FISH<br />
signals than conventional FISH. Consequently, the number of<br />
active rRNA molecules necessary to detect a probe conferred<br />
signal decreases by using CARD FISH (Pernthaler et al.<br />
2002).<br />
DNA array hybridization<br />
DNA array hybridization, also known as Reverse Dot Blot<br />
Hybridization (RDBH) or macroarray, is a technique based<br />
on hybridization of amplified and labelled genome regions<br />
of interest to immobilized oligonucleotides spotted on a<br />
solid support platform. It was originally developed to detect<br />
mutations of human genes, and is still an important diagnostic<br />
tool in clinical research (Chehab & Wall 1992, Yang et al.<br />
2001, Zhang et al. 1991). It is now considered a powerful<br />
and practical technique for the detection and identification<br />
of fungi and other microbes, such as bacteria, from complex<br />
environmental samples without the need for isolation in<br />
culture (Chen et al., 2009, Ehrmann et al. 1994, Lévesque et<br />
al. 1998, Tambong et al. 2006, Uehara et al. 1999, Wu et al.<br />
2007, Zhang et al. 2007, 2008). For this type of application,<br />
oligonucleotides, or microcodes (Summerbell et al. 2005), are<br />
designed from a taxonomically complete dataset of suitable<br />
genome region(s) (Chen et al. 2009, Tambong et al. 2006).<br />
The oligonucleotides can be selected manually, by analysing<br />
multi-sequence alignments, or through computer programs,<br />
such as SigOli (Zahariev et al. 2009) and Array Designer<br />
(Premier Biosoft International, Palo Alto, CA). Synthesized<br />
oligonucleotides with 5’-amine modifications are then spotted<br />
onto a supporting platform, such as a nylon membrane or<br />
glass slide, either manually or robotically. Robotic spotting<br />
can significantly increase the maximum density of the array<br />
which can favour the detection of broader taxonomic groups<br />
(Chen et al. 2009). Amplicons of the target gene region(s)<br />
are amplified by PCR, labelled with digoxygenin (DIG) and<br />
subjected to the DNA hybridization procedure previously<br />
described (Fessehaie et al. 2003). A positive reaction between<br />
an amplicon and a perfectly matched (PM) oligonucleotide<br />
generates a chemiluminescent signal which can be detected<br />
by X-ray film or a digital camera in dark rooms. Captured<br />
images are then analysed on a computer program such as<br />
GenePix Pro (Molecular Devices, Sunnyvale, CA).<br />
The design of species or group-specific oligonucleotides<br />
is a crucial step in this process since it defines the specificity<br />
and sensitivity of the assay (Lievens et al. 2006, Pozhitkov<br />
et al. 2006, Urakawa et al. 2003). It is generally agreed<br />
that the length of the oligonucleotide, the number, type and<br />
position of SNPs contained in an oligonucleotide, determine<br />
its discriminatory potential for DNA hybridization. A desirable<br />
oligonucleotide should: (1) have suitable thermodynamic<br />
characters such as melting temperature; (2) contain as<br />
many polymorphic sites as possible located close to the<br />
centre or on the 3’-half of the sequence; and (3) have the<br />
least probability of hairpin or dimer formation (Kawasaki et al.<br />
1993, Lievens et al. 2006). Oligonucleotide lengths ranging<br />
from 25 to 35 mer have displayed the best balance between<br />
specificity and sensitivity (Chen et al. 2009). Longer lengths<br />
(> 35 mer) of oligonucleotides reduce the ratio of mismatched<br />
to matched base pairs, yet increase the number of bases<br />
available for hybridization, providing lower specificity but<br />
higher sensitivity to an array (Dorris et al. 2003, Lievens et al.<br />
2006). Oligonucleotides up to 70 mer, however, have been<br />
used in macroarrays for the detection of plant viruses using<br />
gene sequences that are widely different between the closely<br />
related pathogens (Agindotan & Perry 2007, 2008). It has<br />
also been demonstrated that more complicated parameters,<br />
such as actual sequence arrangement and the mismatched<br />
duplex type, and their interactions, can play important roles<br />
180<br />
<br />
ima fUNGUS
Molecular identification and detection of fungi<br />
ARTICLE<br />
Fig. 2. DNA macroarray hybridization results from frozen cranberry fruit sample collected from Massachusetts, USA (adapted from Robideau<br />
et al. 2008). The x-ray film is overlaid with the oligonucleotide spotting pattern on DNA array membrane for cranberry fruit rot fungi. The top<br />
left pair of spots and bottom right pair of spots are oligonucleotides which served as positive controls. This array is showing positive signal for<br />
Phyllosticta, Coleophoma, Epicoccum, Godronia, Alternaria, Pestalotia, and Pilidium.<br />
in affecting the discriminatory power of an oligonucleotide<br />
(Urakawa et al. 2003, Lievens et al. 2006, Pozhitkov et al.<br />
2006). Despite numerous attempts to predict the behaviour<br />
of oligonucleotides in DNA hybridization, the thermodynamic<br />
interaction between the probes and oligonucleotides remains<br />
poorly understood (Pozhitkov et al. 2007).<br />
Most DNA macroarrays that have been developed are<br />
based on a single region for the detection of a specific<br />
taxonomic group. Among these regions, 16S ribosomal DNA<br />
has been used for the detection of bacteria (Fessehaie et<br />
al. 2003, Xiong et al. 2006). Various genome regions, such<br />
as ribosomal DNA spacers (ITS), mitochondrial genes (e.g.<br />
cytochrome oxydase c subunit 1, cox1) and some protein<br />
coding regions (β-tubulin, EF-1α, etc), were chosen to target<br />
fungi and fungus-like organisms (Chen et al. 2009, Gilbert et<br />
al. 2008, Harper et al. 2011, Ning et al. 2007, 2008, Seifert<br />
& Lévesque 2004). Oligonucleotides with higher specificity<br />
are often designed from polymorphic sites located at indels<br />
present in multi-sequence alignments (Robideau et al. 2008,<br />
Tambong et al. 2006). Oligonucleotides extracted from a<br />
locus that can be well aligned with low sequence divergence<br />
tends to cross-react with non-target amplicons and display<br />
strong false positive signals even when tested with pure<br />
cultures because of the low frequency of polymorphisms<br />
in the specific oligonucleotides. This has been observed in<br />
arrays using cox1 in Penicillium subgen. Penicillium (Chen<br />
et al. 2009, Seifert et al. 2007). In a recent study, DNA<br />
arrays were constructed from multiple loci of Phytophthora<br />
species, including ITS, cox1 and the intergenic region (cox2-<br />
1 spacer, CS) between cytochrome c oxidase 2 (cox2) and<br />
cox1 (W. Chen et al. pers. comm.). In comparison to the<br />
cox1 region, the length variation and the presence of indels<br />
in the sequence alignments of ITS and CS provided better<br />
opportunities to select highly specific oligonucleotides. The<br />
combination of all three arrays increased the discrimination<br />
potential, detection accuracy, and redundancy of the assay.<br />
DNA arrays have been developed for the detection of<br />
plant pathogens in a wide range of environmental samples,<br />
such as greenhouse crops (Le Floch et al. 2007, Lievens et<br />
al. 2003), potatoes (Fessehaie et al. 2003), ginseng (Punja<br />
et al. 2007), and fruits (Robideau et al. 2008, Sholberg et al.<br />
2005, 2006). Macroarrays are also effective diagnostic tools<br />
for the detection of phytopathogenic bacteria (Fessehaie et<br />
al. 2003), fungi and fungus-like organisms (Chen et al. 2009,<br />
Lévesque et al. 1998, Tambong et al. 2006), nematodes<br />
(Uehara et al. 1999), and viruses (Agindotan & Perry 2007,<br />
2008).<br />
DNA array hybridization is highly sensitive as are most<br />
PCR-based approaches. With the unlimited capacity for<br />
the accommodation of oligonucleotides on one membrane<br />
and the reusability of the membranes, it shows superior<br />
multiplexing detection capability at a lower cost over other<br />
PCR-based methods. In a biodiversity study, the species<br />
profile could be revealed by hybridizing the oligonucleotide<br />
array with a mixed pool of DIG-labelled PCR products<br />
amplified from the total DNA of a query sample. This assay<br />
would be especially valuable for the simultaneous detection<br />
of multiple plant pathogens which cover a broad taxonomic<br />
range but are specific to a certain host. As an example,<br />
Robideau et al. (2008) developed a DNA array for cranberry<br />
fruit rot (CFR) fungal pathogens with over 2000 field samples<br />
being tested. This array had the ability to detect species from<br />
24 genera of fungi known on cranberry with a single test.<br />
Fig. 2 shows that this DNA macroarray was able to detect<br />
species of Phyllosticta, Cladosporium, Epicoccum, Godronia,<br />
Alternaria, Pestalotia, and Pilidium from a single frozen<br />
cranberry fruit sample (Robideau et al. 2008).<br />
The macroarray technology is now available commercially<br />
in four European countries under the name DNA Multiscan<br />
(http://www.dnamultiscan.com) and at the Guelph Pest<br />
Diagnostic Clinic (http://www.guelphlabservices.com/) for<br />
the test of plant pathogens in greenhouse nutrient solutions<br />
and roots of crops. There are very significant advances in next<br />
generation sequencing technology that have reduced the need<br />
of DNA array technology in functional genomics. However,<br />
there are developments in lab-on-a-chip array technology for<br />
very quick and for large-scale testing. A recent example is<br />
a low density array for Phytophthora detection, where amino-<br />
volume 2 · no. 2<br />
181
Tsui et al.<br />
ARTICLE<br />
Fig. 3. Schematic representation of the multiplex tandem–PCR<br />
procedure illustrating specific detection and identification from a blood<br />
sample containing Candida albicans and C. glabrata. Reproduced<br />
from Lau et al. (2009) with the permission of Future Medicine.<br />
labelled oligonucleotides are spotted over a gap between two<br />
electrodes inside a microchip (Julich et al. 2011). Hybridization<br />
is detected by a current passing through a silver deposit over<br />
the gap. Because of such developments, array-based assays<br />
are likely to remain a popular option for diagnostics.<br />
Multiplex tandem PCR<br />
Multiplexed-tandem PCR (MT-PCR) is a technology platform<br />
developed for highly multiplexed gene expression profiling<br />
and the rapid identification of clinically important pathogens<br />
(Stanley et al. 2005). The platform consists of two rounds<br />
of amplification (Fig. 3). In the first step, a multiplex PCR is<br />
performed at 10 to 15 cycles to allow enrichment of target<br />
DNA without creating competition between amplicons (Lau<br />
et al. 2009). This product is diluted and used as template for<br />
the second amplification that consists of multiple individual<br />
quantitative PCR reactions with primers nested within<br />
those used in the multiplex PCR. Up to 72 different PCR<br />
reactions can be multiplexed and performed simultaneously.<br />
Fluorescence is measured by SYBR green technology at<br />
the end of each extension cycle, and melt-curve analysis<br />
provides species-specific or gene-specific identification.<br />
The incorporation of two sets of species-specific primers for<br />
each target ensures correct amplification and detection, thus<br />
avoiding the expense of DNA probes. SYBR green detection<br />
also increases the multiplexing and quantitative capacity<br />
of real-time PCR systems, which are usually limited by the<br />
availability of fluorescent channels and the need to optimize<br />
each individual multiplex PCR.<br />
In the clinical setting, invasive fungal infections (IFIs)<br />
remain a leading cause of morbidity and mortality in<br />
immune-compromised hosts. MT-PCR was considered a<br />
suitable alternative for the rapid detection and identification<br />
of fungal pathogens directly from clinical specimens, thus<br />
circumventing the need for gold standard culture and<br />
histology, which is slow, insensitive and non-specific. Faster<br />
and accurate molecular identification would also enable<br />
better guidance and earlier administration of targeted<br />
antifungal therapies. As such, the laboratory at Westmead<br />
hospital, in conjunction with AusDiagnostics (Alexandria,<br />
NSW, Australia), developed several MT-PCR assays to<br />
detect the16 major causes of fungal bloodstream infections.<br />
The targets included 11 species of Candida, Cryptococcus<br />
neoformans complex, Saccharomyces cerevisiae, pan-<br />
Fusarium, F. solani, and Scedosporium prolificans. Primers<br />
were designed using sequence variations within the ITS<br />
regions, elongation factor 1-a (EF1-a), and b-tubulin genes.<br />
Fungal targets were selected according to their frequency<br />
of causing infections, their potential resistance to frontline<br />
antifungal agents (especially fluconazole), and their high<br />
attributable mortality.<br />
The MT-PCR platform was initially evaluated on 70 blood<br />
cultures in which a yeast or mould was seen in Gram’s stain<br />
preparations, as well as 200 bacterial blood cultures and 30<br />
samples which did not flag positive (Lau et al. 2008a). The<br />
sensitivity and specificity of the assay was 100 %. This included<br />
the correct identification of fungi in five cases with bacterial<br />
co-infection. In addition, no interference was observed in<br />
simulated cases of polyfungal infection. Unfortunately, three<br />
rare disease-causing yeasts (Candida lambica, C. nivariensis<br />
and Kodamaea ohmeri) were not detected by MT-PCR as<br />
targets were not available on the fungal panel. Nevertheless,<br />
this study demonstrated the diagnostic usefulness of the<br />
platform to rapidly identify common fungal pathogens within<br />
four hours of blood culture flagging (including automated<br />
nucleic acid extraction), which is considerably faster than the<br />
48-96 h required by gold standard methods.<br />
An expanded version of the MT-PCR fungal platform was<br />
then evaluated on 255 EDTA whole blood, 29 serum, and 24<br />
plasma specimens obtained from 109 patients with proven<br />
candidemia (Lau et al. 2010) with the aim of circumventing<br />
the technical and sensitivity <strong>issue</strong>s inherent in routine blood<br />
culture diagnosis. Although the MT-PCR assay was less<br />
sensitive than blood culture (75% sensitivity), the diagnosis<br />
of candidemia and pathogen identification was expedited by<br />
up to four days. The results also supported the contention<br />
that serum and plasma samples were better than whole<br />
blood samples for the molecular detection of candidemia.<br />
Using a technique known as colony MT-PCR (Lau et<br />
al. 2008b), the fungal assay was also able to provide rapid<br />
(1.5 h) and unambiguous identification of yeasts direct from<br />
primary isolation plates without the need for DNA extraction or<br />
separation from mixed fungal/bacterial cultures. As such, colony<br />
MT-PCR offered a faster and better alternative to biochemical<br />
assays which are often subjective, prone to misidentification,<br />
and dependent on a pure culture being obtained. Current work<br />
is aimed at integrating identification panels with targets to<br />
detect molecular mechanisms of antifungal resistance.<br />
Overall, the fungal MT-PCR assay compares favourably<br />
to other commercial platforms and integrates well into the<br />
182<br />
<br />
ima fUNGUS
Molecular identification and detection of fungi<br />
ARTICLE<br />
Fig. 4. Schematic overview of the padlock principle combined with OpenArray ® , Technology for multiplex detection of three different targets<br />
(adapted from van Doorn et al. 2007).<br />
routine workflow of diagnostic laboratories. Automated<br />
operations and use of commercial reagents further enables<br />
standardized procedures to be established. Nevertheless, the<br />
major limitation of all molecular diagnostic assays is target<br />
availability, which is often dictated by an effort to balance out<br />
costs and turn-around time with maximum throughput. The<br />
increased sensitivity of molecular assays and its ability to<br />
detect viable and non-viable cells should also place heavy<br />
emphasis on interpreting results with other microbiological<br />
data and clinical information.<br />
ISOTHERMAL SYSTEMS<br />
Isothermal systems do not require a thermal cycler to produce<br />
rapid temperature changes, but require only a simple platform<br />
such as heating blocks or water bath. Isothermal systems<br />
include rolling circle amplification (RCA) and loop-mediated<br />
isothermal amplification (LAMP).<br />
Padlock probe technology and rolling circle<br />
amplification<br />
Detection and characterization of single nucleotide<br />
polymorphisms (SNPs) is becoming increasing popular<br />
for pathogen identification, but was considered a major<br />
challenge for conventional real-time PCR using regular<br />
oligonucleotides detected by fluorescent dyes (e.g. SYBR<br />
green or TaqMan probes). In order to recognize SNPs among<br />
different genotypes, padlock probe techniques are required.<br />
Padlock probes (PLPs) are long oligonucleotides (about 100<br />
bp) carrying a non-target-complementary segment flanked<br />
by the target complementary regions at their 5’ and 3’ ends,<br />
which recognize adjacent sequences on the target DNA<br />
(Nilsson et al. 1994). Thus, on hybridization, the ends of<br />
the probes occupy adjacent positions, and can be joined by<br />
enzymatic ligation (Figs 4–5). Ligation occurs and the probes<br />
are circularized only when both end segments correctly<br />
recognize the target sequences (Landegren et al. 1988). The<br />
helical nature of double-stranded DNA (dsDNA) enables the<br />
volume 2 · no. 2<br />
183
Tsui et al.<br />
ARTICLE<br />
Fig. 5. Schematic representation of the steps in padlock probe<br />
technology coupled with hyperbranched rolling circle amplification<br />
(H-RCA) for SNPs detection. 1. The hybridization of padlock probes<br />
(containing the complementary sequences at the 5’ and 3’ ends) to<br />
the target templates. 2. During a perfect match, the probe forms a<br />
circular molecular with the aid of DNA ligase; while in the case of<br />
mismatch, no circular molecules formed. 3. Non-hybridized template<br />
will be removed during the exonucleolysis reactions (digestion<br />
by exonucleases I and III). 4. H-RCA is performed using two predesigned<br />
primers and Bst polymerase, but no amplification will take<br />
place in the absence of a circular molecular. 5. The accumulation of<br />
dsDNA products during isothermal rolling circle amplification of DNA<br />
minicircles is monitored in a real time PCR thermocycler with the<br />
addition of SYBR green.<br />
probe to topologically bind to the target strand and the probe<br />
can’t be displaced (Nilsson et al. 1994).<br />
Padlock probes were initially introduced for in situ<br />
DNA localization and detection (Nilsson et al. 1994). They<br />
were developed originally for discrimination of centromeric<br />
sequence variation in human chromosomes (Nilsson et<br />
al. 1997). However, the method has now been applied to<br />
detection of genetically modified organisms (Prins et al.<br />
2008). This concept also provides extensive multiplex<br />
potential for pathogen detection as the interaction between<br />
padlock probes does not give rise to circular molecules,<br />
which can be easily removed from the detection system<br />
(Nilsson et al. 1994, 1996). Recently, padlock probe-based<br />
applications for multiplex quantitative targets detection and<br />
for genotyping fungal and microbial community analysis<br />
using high throughput real-time PCR on OpenArrays ® have<br />
been developed (van Doorn et al. 2007). Advantages of<br />
PLP-based diagnostic applications developed are a flexible<br />
and easily adaptable design, specificity, and multiplexing,<br />
universal downstream processing after ligation, and highthroughput<br />
format with real-time analysis (Tsui et al. 2012)<br />
(Fig. 4).<br />
Briefly, various padlock probes are designed to target<br />
organisms and ligated to DNA extracted from environmental<br />
samples or cultures (van Doorn et al. 2007). Targets for<br />
ligation present in complex DNA samples such as soil or recirculated<br />
water can be generated by PCR pre-amplification,<br />
through Phi29 polymerase and whole genome amplification to<br />
ensure efficient detection. Real-time quantification for multiple<br />
targets is performed in OpenArrays ® , which can accommodate<br />
up to 3072 x 33 nl PCR amplification with preloaded probespecific<br />
primers (Fig. 4). Multiplex padlock ligation is followed<br />
by single-plex amplification using unique probe-specific primer<br />
pairs and SYBR green based detection in nano-litre PCR<br />
array (van Doorn et al. 2007). The performance of the padlock<br />
probe detection system has been demonstrated using 13<br />
probes targeting several plant pathogens at various taxonomic<br />
levels (Szemes et al. 2005, van Doorn et al. 2007). All<br />
probes specifically detected their corresponding targets, and<br />
provided perfect discrimination against non-target organisms<br />
with very similar target sites. Pathogen quantification was<br />
robust in single target versus mixed target assays. This novel<br />
assay enables very specific, high-throughput, quantitative<br />
detection of multiple pathogens over a wide range of target<br />
concentrations, and should be easily adaptable for versatile<br />
diagnostic purposes (van Doorn et al. 2007).<br />
Also, padlock probes containing zip-code sequence or a<br />
biotin-labelled moiety and internal endonuclease cleavage<br />
site, in conjunction with quantitative PCR and Luminex TM<br />
technology or microarray technology, can be used for<br />
multiplex pathogen detection and quantification (Szemes et<br />
al. 2005, Eriksson et al. 2009, van Doorn et al. 2009).<br />
Alternatively, the signal by which the target hybridizes<br />
perfectly to the padlock probes can be amplified by<br />
hyperbranched rolling circle amplification (H-RCA) (Banér<br />
et al. 1998). Rolling circle amplification (RCA) is based on<br />
the rolling replication of short single stranded DNA (ssDNA)<br />
circular molecules (Lizardi et al. 1998, Fire & Xu 1995). RCA<br />
involves a single forward primer complementary to the linker<br />
region of the padlock probe and a DNA polymerase with strand<br />
displacement activity in an isothermal condition (Pickering<br />
et al. 2002). As a result, the padlock probe signal can be<br />
amplified several thousand-fold because the polymerase<br />
extends the bound primer along the padlock probes for many<br />
cycles and displaces upstream sequences, producing a long<br />
ssDNA molecule comprising multiple repeats of the probe<br />
sequence. Two primers are employed: a first forward primer<br />
that binds to the padlock probe and initializes RCA, and a<br />
184<br />
<br />
ima fUNGUS
Molecular identification and detection of fungi<br />
Fig. 6. Schematic representation of the mechanism of LAMP. A.<br />
General location of the LAMP primer set in relation to defined regions<br />
of the target DNA. Forward (F3) and backward (B3) outer primers<br />
and forward (FIP) and backward (BIP) inner primers are indicated.<br />
B, C. Basic principle and amplification steps in LAMP. In general<br />
new DNA strands are synthesizes from the F3 and B3 primers, and<br />
these strands are recognized by FIP and BIP to start loop-mediated<br />
autocycling amplifications. The final products are stem-loop DNAs<br />
with several inverted repeats of the target DNA, and cauliflower-like<br />
structures bearing multiple loops (modified from diagrams at © Eiken Chemical.<br />
second primer that targets the repeated ssDNA sequence of<br />
the primary RCA product, finally generating large numbers of<br />
copies of the DNA fragments. This is called hyperbranching<br />
RCA (H-RCA) (Lizardi et al. 1998) (Fig. 5).<br />
Padlock probe coupled with H-RCA offers a significant<br />
advantage for the detection of SNPs (Tsui et al. 2012). The<br />
formation of circular probes via ligation occurs when both<br />
ends of the padlock probes perfectly hybridize to the target at<br />
juxtaposition (Fig. 5). The subsequent H-RCA amplification of<br />
a target probe could be carried out when circularized probes<br />
become available. These two strict conditions create an ideal<br />
detection platform for highly sensitive and specific SNPs<br />
detection. By increasing the hybridization temperature and<br />
shortening the 3’ arm (below the reaction temperature), the<br />
discrimination of SNP can be further improved (Pickering et<br />
al. 2002, Faruqi et al. 2001). This method for SNPs detection<br />
based on DNA ligase-mediated single nucleotide discrimination<br />
with consecutive signal amplification by H-RCA has been<br />
developed for various groups of pathogenic organisms,<br />
including fungi, bacteria, and viruses (Kong et al. 2008, Zhou<br />
et al. 2008, Kaocharoen et al. 2008, Wang et al. 2009, 2010).<br />
Recently, the technology has been used to differentiate and to<br />
detect two closely related conifer pathogens vectored by the<br />
mountain pine beetles (Tsui et al. 2010).<br />
Loop mediated isothermal amplification<br />
Loop-mediated isothermal amplification (LAMP) is a powerful<br />
and novel nucleic acid amplification method that amplifies<br />
a few copies of target DNA with high specificity, efficiency,<br />
and rapidity under isothermal conditions, using a set of four<br />
specially designed primers and a DNA polymerase with strand<br />
displacement activity (Notomi et al. 2000, Parida et al. 2008,<br />
Tomita et al. 2008). The cycling reactions can result in the<br />
accumulation of 10 9 to 10 10 -fold copies of target in less than<br />
an hour. Considering the advantages of rapid amplification,<br />
simple operation and easy detection, LAMP has potential<br />
applications for clinical diagnosis as well as surveillance of<br />
infectious diseases in developing countries without requiring<br />
sophisticated equipment or skilled personnel (Mori & Notomi<br />
2009, Parida et al. 2008).<br />
The technique was first described and initially evaluated<br />
for detection of hepatitis B virus DNA by Notami et al.<br />
(Notomi et al. 2000). LAMP assays have been mostly used<br />
for the diagnostics of bacteria (Chen et al. 2011, Han et<br />
al. 2011), virus (Wang et al. 2011, Zhao et al. 2011), and<br />
parasites (Ikadai et al. 2004, Iseki et al. 2007), but it has<br />
also been developed for the rapid detection of pathogenic<br />
or allergenic fungi. Ohori et al. (2006) used the technique for<br />
rapid identification of Ochroconis gallopava, an emerging<br />
fungal pathogen and causative agent of zoonotic infections,<br />
while Sun and co-workers, used it for the rapid diagnosis of<br />
Penicillium marneffei in archived t<strong>issue</strong> samples (Sun et al.<br />
2010a) and of Fonsecaea agents of chromoblastomycosis<br />
(Sun et al. 2010b). Similarly Endo et al. (2004) and Tatibana<br />
et al. (2009) detected the presence of the gp43 gene in<br />
Paracoccidioides brasiliensis by LAMP, and Lucas et al.<br />
(2010) used LAMP for differentiation of Cryptococcus<br />
neoformans varieties from C. gattii based on CAP59<br />
sequences. Recently, Lu et al. (2011) utilized the technology<br />
for the identification of Pseudallescheria and Scedosporium<br />
species.<br />
Two inner and two outer primers are required for LAMP<br />
(Fig. 6A). In the initial steps of the LAMP reaction, all four<br />
primers are employed, but in the later cycling steps only<br />
the inner primers are used for strand displacement DNA<br />
synthesis. The outer primers are referred to as F3 and B3,<br />
while the inner primers are forward inner primer (FIB) and<br />
backward inner primer (BIP). Both FIP and BIP contains two<br />
distinct sequences corresponding to the sense and antisense<br />
sequences of the target DNA, one for priming in the first<br />
stage and the other for self-priming in later stages (Notomi et<br />
al. 2000). The size and sequence of the primers was chosen<br />
so that their melting temperature (Tm) is between 60-65 °C,<br />
the optimal temperature for Bst polymerase. The F1c and<br />
B1c Tm values should be a little higher than those of F2 and<br />
B2 to form the looped-out structure. The Tm values of the<br />
outer primers F3 and B3 have to be lower than those of F2<br />
and B2 to assure that the inner primers start synthesis earlier<br />
than the outer primers. Additionally, the concentrations of the<br />
inner primers are higher than the concentrations of the outer<br />
primers (Notomi et al. 2000, Tomita et al. 2008). Furthermore,<br />
it is critical for LAMP to form a stem-loop DNA from a dumb-<br />
ARTICLE<br />
volume 2 · no. 2<br />
185
Tsui et al.<br />
ARTICLE<br />
bell structure. Various sizes of the loop between F2c and<br />
F1c and between B2c and B1c were examined, and the best<br />
results are obtained when loops of 40 nucleotides (40nt)<br />
or longer are used (Notomi et al. 2000). The size of target<br />
DNA is an important factor that LAMP efficiency depends<br />
on, because the rate limiting step for amplification is strand<br />
displacement DNA synthesis.<br />
LAMP relies on auto-cycling strand displacement DNA<br />
synthesis in the presence of Bst DNA polymerase, specific<br />
primers and the target DNA template. The mechanism of<br />
the LAMP amplification reaction includes three steps:<br />
production of starting material, cycling amplification, and<br />
recycling (Notomi et al. 2000, Tomita et al. 2008) (Fig. 6B, C).<br />
To produce the starting material, inner primer FIB hybridizes<br />
to F2c in the target DNA and initiates complementary strand<br />
synthesis. Outer primer F3 hybridizes to F3c in the target<br />
and initiates strand displacement DNA synthesis, releasing<br />
a FIP-linked complementary strand, which forms a loopedout<br />
structure at one end (DNA amplification proceeds with<br />
BIP in a similar manner). This single stranded DNA serves<br />
as template for BIP-initiated DNA synthesis and subsequent<br />
B3-primed strand displacement DNA synthesis leading to<br />
the production of a dumb-bell form DNA which is quickly<br />
converted to a stem loop DNA. This then serves as the<br />
starting material for LAMP cycling, the second stage of the<br />
LAMP reaction. During cycling amplification, FIP hybridizes<br />
to the loop in the stem-loop DNA and primes strand<br />
displacement DNA synthesis, generating as an intermediate<br />
one gapped stem loop DNA with an additional inverted copy<br />
of the target sequence in the stem, and a loop formed at<br />
the opposite end via the BIP sequence. Subsequent selfprimed<br />
strand displacement DNA synthesis yields one<br />
complementary structure of the original stem-loop DNA, and<br />
one gap repaired stem-loop DNA with a stem elongated to<br />
twice as long and a loop at the opposite end. Both of these<br />
products then serve as templates for BIP-primed strand<br />
displacement in the subsequent cycles, the elongation and<br />
recycling step. The final product is a mixture of stem loop<br />
DNA with various stem length and cauliflower-like structures<br />
with multiple loops formed by annealing between alternately<br />
inverted repeats of the target sequence in the same strand<br />
(Notomi et al. 2000, Tomita et al. 2008).<br />
LAMP products can be directly observed by the naked eye<br />
or using a UV transilluminator in the reaction tube by adding<br />
2.0 µl of 10 fold diluted SYBR Green I stain to the reaction<br />
tube separately. Under UV illumination, the gel shows a<br />
ladder-like structure from the minimum length of target DNA<br />
up to the loading well, which are the various length stem-loop<br />
products of the LAMP reaction.<br />
Conclusion<br />
Numerous detection methodologies are now available, but<br />
regardless of the approach, important questions need to be<br />
answered prior to their inclusion into experiments. These<br />
include sensitivity, accuracy, robustness, frequency of testing,<br />
and cost. Despite many novel technologies being available,<br />
challenges remain to identify as yet unculturable fungi, to<br />
detect cryptic species, and to characterize the assemblage<br />
and diversity of fungal communities in different environments<br />
without bias. There is always a need to characterize fungi<br />
quickly and accurately. No one knows how many fungal<br />
species exist, but sequencing of environmental DNA may<br />
improve the accuracy of current estimates (Hawksworth<br />
2001). Next-generation sequencing and pyrosequencing<br />
approaches will also provide promising ways of enlarging the<br />
scope of molecular-detection studies.<br />
Acknowledgements<br />
We would like to thank the IMC9 organizers for supporting this SIG<br />
symposium. We are thankful to Satoko Yamamoto (Eiken Chemical,<br />
Japan) for permission to use diagrams of LAMP from .<br />
References<br />
Agindotan B, Perry KL (2007) Macroarray detection of plant RNA<br />
viruses using randomly primed and amplified complementary<br />
DNAs from infected plants. Phytopathology 97: 119–127.<br />
Agindotan B, Perry KL. (2008) Macroarray detection of eleven<br />
potato-infecting viruses and Potato spindle tuber viroid. Plant<br />
Disease 92: 730–740.<br />
Agrios G (2005) Plant Pathology. 5 th edn. London: Elsevier Academic<br />
Press.<br />
Amann RI, Ludwig W and Schleifer K-H (1995) Phylogenetic<br />
identification and in situ detection of individual microbial cells<br />
without cultivation. Microbiology and Molecular Biology Reviews<br />
59: 143–169.<br />
Baker BJ, Lutz MA, Dawson SC, Bond PL, Banfield JF (2004)<br />
Metabolically active eukaryotic communities in extremely acidic<br />
mine drainage. Applied and Environmental Microbiology 70:<br />
6264–6271.<br />
Banér J, Nilsson M, Mendel-Hartvig M, Landegren U (1998) Signal<br />
amplification of padlock probes by rolling circle replication.<br />
Nucleic Acids Research 26: 5073–5078.<br />
Baschien C, Manz W, Neu TR, Szewzyk U (2001) Fluorescence in<br />
situ hybridization of freshwater fungi. International Review of<br />
Hydrobiology 86: 371–384.<br />
Baschien C, Manz W, Neu TR, Marvanová L, Szewzyk U (2008) In<br />
situ detection of freshwater fungi in an alpine stream by new<br />
taxon-specific fluorescence in situ hybridization probes. Applied<br />
and Environmental Microbiology 74: 6427–6436.<br />
Brierley JL, Stewart JA, Lees AK (2009) Quantifying potato pathogen<br />
DNA in soil. Applied Soil Ecology 41: 234–238.<br />
Brul S, Nussbaum J, Dielbandhoesing SK (1997) Fluorescent probes<br />
for cell wall 16 porosity and membrane integrity in filamentous<br />
fungi. Journal of Microbiological Methods 28: 169–178.<br />
Chehab FF, Jeff W (1992) Detection of multiple cystic fibrosis<br />
mutations by reverse dot blot hybridization: a technology for<br />
carrier screening. Human Genetics 89: 163–168.<br />
186<br />
<br />
ima fUNGUS
Molecular identification and detection of fungi<br />
Chen J, Ma X, Yuan Y, Zhang W (2011) Sensitive and rapid detection<br />
of Alicyclobacillus acidoterrestris using loop-mediated isothermal<br />
amplification. Journal of the Sciemce of Food and Agriculture 91:<br />
1070–1074.<br />
Chen W, Seifert K, Lévesque CA (2009) A high density COX1<br />
barcode oligonucleotide array for identification and detection of<br />
species of Penicillium subgenus Penicillium. Molecular Ecology<br />
Resources 9: 114–129.<br />
Endo S, Komori T, Ricci G, Sano A, Yokoyama K, Ohori A, Kamei<br />
K, Franco M, Miyaji M, Nishimura K (2004) Detection of gp43<br />
of Paracoccidioides brasiliensis by the loop-mediated isothermal<br />
amplification (LAMP) method. FEMS Microbiology Letters 234:<br />
93–97.<br />
Eriksson R, Jobs M, Ekstrand C, Ullberg M, Herrmann B,<br />
Landegren U, Nilsson M, Blomberg J (2009) Multiplex and<br />
quantifiable detection of nucleic acid from pathogenic fungi<br />
using padlock probes, generic real time PCR and specific<br />
suspension array readout. Journal of Microbiological Methods<br />
78: 195–202.<br />
Faruqi AF, Hosono S, Driscoll MD, Dean FB, Alsmadi O, Bandaru<br />
R, Kumar G, Grimwade B, Zong Q, Sun Z, Du Y, Kingsmore S,<br />
Knott T, Lasken RS (2001) High-throughput genotyping of single<br />
nucleotide polymorphisms with rolling circle amplification. BMC<br />
Genomics 2: 4.<br />
Fessehaie A De Boer SH, Lévesque CA (2003) An oligonucleotide<br />
array for the identification and differentiation of bacteria<br />
pathogenic on potato. Phytopathology 93: 262–269.<br />
Fire A, Xu SQ (1995) Rolling replication of short DNA circles.<br />
Proceedings of the National Academy of Sciences, USA 92:<br />
4641–4645.<br />
Gilbert CA, Zhang N, Hutmacher RB, Davis RM, Smart CD (2008)<br />
Development of a DNA-based macroarray for the detection and<br />
identification of Fusarium oxysporum f. sp. vasinfectum in cotton<br />
t<strong>issue</strong>. Journal of Cotton Science 12: 165–170.<br />
Han HJ, Jung SJ, Oh MJ, Kim DH (2011) Rapid and sensitive<br />
detection of Streptococcus iniae by loop-mediated isothermal<br />
amplification (LAMP). Journal of Fish Disease 34: 395–8.<br />
Harper KA, Smart CD, Michael Davis R (2011) Development of a<br />
DNA-based macroarray for the detection and identification of<br />
Amanita species. Journal of Forensic Science 56: 1003–1009.<br />
Hawksworth DL (1991) The fungal dimension of biodiversity:<br />
magnitude, significance, and conservation. Mycological<br />
Research 95: 641–655.<br />
Hawksworth DL (2001) The magnitude of fungal diversity: the 1.5<br />
million species estimate revisted. Mycological Research 105:<br />
1422–1432.<br />
Ikadai H, Tanaka H, Shibahara N, Matsuu A, Uechi M, Itoh N,<br />
Oshiro S, Kudo N, Igarashi I, Oyamada T (2004) Molecular<br />
evidence of infections with Babesia gibsoni parasites in Japan<br />
and evaluation of the diagnostic potential of a loop-mediated<br />
isothermal amplification method. Journal of Clinical Microbiology<br />
42: 2465–2469.<br />
Inácio J, Behrens S, Fuchs BM, Fonseca Á, Spencer-Martins I, Amann<br />
R (2003) In situ accessibility of Saccharomyces cerevisiae 26S<br />
rRNA to cy3-labeled oligonucleotide probes comprising the D1<br />
and D2 domains. Applied and Environmental Microbiology 69:<br />
2899–2905.<br />
Iseki H, Alhassan A, Ohta N, Thekisoe OM, Yokoyama N, Inoue<br />
N, Nambota A, Yasuda J, Igarashi I (2007) Development of a<br />
multiplex loop-mediated isothermal amplification (mLAMP)<br />
method for the simultaneous detection of bovine Babesia<br />
parasites. Journal of Microbiological Methods 71: 281–287.<br />
Julich S, Riedel M, Kielpinski M, Urban M, Kretschmer R, Wagner S,<br />
Fritzsche W, Henkel T, Möller R, Werres S (2011) Development<br />
of a lab-on-a-chip device for diagnosis of plant pathogens.<br />
Biosensors and Bioelectronics 26: 4070–4075.<br />
Kaocharoen S, Wang B, Tsui KM, Trilles L, Kong F, Meyer W (2008)<br />
Hyperbranched rolling circle amplification as a rapid and sensitive<br />
method for species identification within the Cryptococcus species<br />
complex. Electrophoresis 29: 3183–3191.<br />
Kawasaki E, Saiki R, Erlich H (1993) Genetic analysis using<br />
polymerase chain reaction-amplified DNA and immobilized<br />
oligonucleotide probes: reverse dot-blot typing. Methods in<br />
Enzymology 218: 369–381.<br />
Kong F, Tong Z, Chen X, Sorrell T, Wang B, Wu Q, Ellis D, Chen<br />
SC (2008) Rapid identification and differentiation of Trichophyton<br />
species, based on sequence polymorphisms of the ribosomal<br />
internal transcribed spacer regions, by rolling-circle amplification.<br />
Journal of Clinical Microbiology 46: 1192–1199.<br />
Landegren U, Kaiser R, Sanders J, Hood L (1988) A ligase-mediated<br />
gene detection technique. Science 241: 1077–1080.<br />
Lau A, Chen S, Sleiman S, Sorrell T (2009) Current status and future<br />
perspectives on molecular and serological methods in diagnostic<br />
mycology. Future Microbiology 4: 1185–1222.<br />
Lau A, Sorrell TC, Chen S, Stanley K, Iredell J, Halliday C (2008a)<br />
Multiplex tandem PCR: a novel platform for rapid detection and<br />
identification of fungal pathogens from blood culture specimens.<br />
Journal of Clinical Microbiology 46: 3021–3027.<br />
Lau A, Sorrell TC, Lee O, Stanley K, Halliday C (2008b) Colony<br />
multiplex-tandem PCR for rapid, accurate identification of fungal<br />
cultures. Journal of Clinical Microbiology 46: 4058–4060.<br />
Lau A, Halliday C, Chen SCA, Playford EG, Stanley K, Sorrell TC<br />
(2010) Comparison of whole blood, serum and plasma for early<br />
detection of candidemia by multiplex-tandem PCR. Journal of<br />
Clinical Microbiology 48: 811–816.<br />
Le Floch G, Tambong J, Vallance J, Tirilly Y, Lévesque A, Rey P<br />
(2007) Rhizosphere persistence of three Pythium oligandrum<br />
strains in tomato soilless culture assessed by DNA macroarray<br />
and real-time PCR. FEMS Microbiology Ecology 61:317–326.<br />
Lévesque CA, Harlton CE, de Cock AWAM (1998) Identification<br />
of some oomycetes by reverse dot blot hybridization.<br />
Phytopathology 88: 213–222.<br />
Li S, Cullen D, Hjort M, Spear R, Andrews JH (1996) Development<br />
of an oligonucleotide probe for Aureobasidium pullulans based<br />
on the small subunit rRNA gene. Applied and Environmental<br />
Microbiology 62:1514–1518.<br />
Lievens B, Brouwer M, Vanachter ACRC, Lévesque CA, Cammue BPA,<br />
Thomma BPHJ (2003) Design and development of a DNA array<br />
for rapid detection and identification of multiple tomato vascular<br />
wilt pathogens. FEMS Microbiology Letters 223: 113–122.<br />
Lievens B, Claes L, Vanachter AC, Cammue BP, Thomma BP (2006)<br />
Detecting single nucleotide polymorphisms using DNA arrays<br />
for plant pathogen diagnosis. FEMS Microbiology Letters 255:<br />
129–139.<br />
ARTICLE<br />
volume 2 · no. 2<br />
187
Tsui et al.<br />
ARTICLE<br />
Lizardi PM, Huang X, Zhu Z, Bray-Ward P, Thomas DC, Ward DC<br />
(1998) Mutation detection and single-molecule counting using<br />
isothermal rolling-circle amplification. Nature Genetics 19: 225–<br />
232.<br />
Lu Q, Gerrits van den Ende AH, Bakkers JM, Sun J, Lackner M,<br />
Najafzadeh MJ, Melchers WJ, Li R, de Hoog GS (2011).<br />
Identification of Pseudallescheria and Scedosporium species<br />
by three molecular methods. Journal of Clinical Microbiology 49:<br />
960-7<br />
Lucas S, da Luz Martins M, Flores O, Meyer W, Spencer-Martins<br />
I, Inácio J (2010) Differentiation of Cryptococcus neoformans<br />
varieties and Cryptococcus gattii using CAP59-based loopmediated<br />
isothermal DNA amplification. Clinical Microbiology<br />
and Infection 16: 711–714.<br />
McArthur FA, Bärlocher MO, McLean NAB, Hiltz MD, Bärlocher<br />
F (2001) Asking probing questions: can fluorescent in situ<br />
hybridization identify and localize aquatic hyphomycetes on leaf<br />
litter? International Review of Hydrobiology 86: 429-438.<br />
Mori Y, Notomi T (2009) Loop-mediated isothermal amplification<br />
(LAMP): a rapid, accurate, and cost-effective diagnostic method<br />
for infectious diseases. Journal of Infection and Chemotherapy<br />
15: 62–69.<br />
Nilsson M, Malmgren H, Samiotaki M, Kwiatkowski M, Chowdhary<br />
BP, Landegren U (1994) Padlock probes: circularizing<br />
oligonucleotides for localized DNA detection. Science 265:<br />
2085–2088.<br />
Nilsson M, Krejci K, Koch J, Kwiatkowski M, Gustavsson P,<br />
Landegren U (1997) Padlock probes reveal single-nucleotide<br />
differences, parent of origin and in situ distribution of centromeric<br />
sequences in human chromosomes 13 and 21. Nature Genetics<br />
16: 252–255.<br />
Nilsson M, Dahl F, Larsson C, Gullberg M, Stenberg J (2006)<br />
Analyzing genes using closing and replicating circles. Trends in<br />
Biotechnology 24: 83–88.<br />
Notomi T, Okayama H, Masubuchi H, Yonekawa T, Watanabe K,<br />
Amino N, Hase T (2000) Loop-mediated isothermal amplification<br />
of DNA. Nucleic Acids Research 28: E63.<br />
Ohori A, Endo S, Sano A, Yokoyama K, Yarita K, Yamaguchi M, Kamei<br />
K, Miyaji M, Nishimura K (2006) Rapid identification of Ochroconis<br />
gallopava by a loop-mediated isothermal amplification (LAMP)<br />
method. Veterinary Microbiology 114: 359–365.<br />
Okubara PA, Schroeder KL, Li C, Schumacher RT, Lawrence NP<br />
(2007) Improved extraction of Rhizoctonia and Pythium DNA<br />
from wheat roots and soil samples using pressure cycling<br />
technology. Canadian Journal of Plant Pathology 29: 304–<br />
310.<br />
Ophel-Keller K, McKay A, Hartley D, Herdina A, Curran J (2008)<br />
Development of a routine DNA-based testing service for<br />
soilborne diseases in Australia. Australasian Plant Pathology<br />
37: 243–253.<br />
Parida M, Sannarangaiah S, Dash PK, Rao PV, Morita K (2008) Loop<br />
mediated isothermal amplification (LAMP): a new generation of<br />
innovative gene amplification technique; perspectives in clinical<br />
diagnosis of infectious diseases. Reviews in Medical Virology 18:<br />
407–421.<br />
Pernthaler A, Pernthaler J, Amann R (2002) Fluorescence in<br />
situ hybridization and catalyzed reporter deposition for the<br />
identification of marine bacteria. Applied and Environmental<br />
Microbiology 68: 3094–3101.<br />
Pickering J, Bamford A, Godbole V, Briggs J, Scozzafava G, Roe P,<br />
Wheeler C, Ghouze F, Cuss S (2002) Integration of DNA ligation<br />
and rolling circle amplification for the homogeneous, end-point<br />
detection of single nucleotide polymorphisms. Nucleic Acids<br />
Research 30: e60.<br />
Pozhitkov A, Noble PA, Domazet-Losˇo T, Nolte AW, Sonnenberg R,<br />
Staehler P, Beier M, Tautz D (2006) Tests of rRNA hybridization<br />
to microarrays suggest that hybridization characteristics of<br />
oligonucleotide probes for species discrimination cannot be<br />
predicted. Nucleic Acids Research 34: e66.<br />
Pozhitkov AE, Tautz D, Noble PA (2007) Oligonucleotide microarrays:<br />
widely applied – poorly understood. Briefings in Functional<br />
Genomics & Proteomics 6: 141–148.<br />
Prins TW, van Dijk JP, Beenen HG, Van Hoef AA, Voorhuijzen MM,<br />
Schoen CD, Aarts HJ, Kok EJ (2008) Optimised padlock probe<br />
ligation and microarray detection of multiple (non-authorised)<br />
GMOs in a single reaction. BMC Genomics 9: 584.<br />
Punja ZK, Wan A, Goswami RS, Verma N, Rahman M, Barasubiye<br />
T, Seifert KA, Lévesque CA (2007) Diversity of Fusarium species<br />
associated with discolored ginseng roots in British Columbia.<br />
Canadian Journal of Plant Pathology 29: 340–353.<br />
Reeleder RD, Capella BB, Tomlinson LD, Hickey WJ (2003) The<br />
extraction of fungal DNA from multiple large soil samples.<br />
Canadian Journal of Plant Pathology 25: 182–191.<br />
Robideau GP, Caruso FL, Oudemans PV, McManus PS, Renaud MA,<br />
Auclair ME, Bilodeau GJ, Yee D, Désaulniers NL, DeVerna JW,<br />
Lévesque CA (2008) Detection of cranberry fruit rot fungi using<br />
DNA array hybridization. Canadian Journal of Plant Pathology<br />
30: 226–240.<br />
Robin BJ, Arffa RC, Avni I, Rao NA (1986) Rapid visualization of three<br />
common fungi using fluorescein-conjugated lectins. Investigative<br />
Ophthalmology & Visual Science 27: 500–506.<br />
Scupham AJ, Presley LL, Wei B, Bent E, Griffith N, McPherson<br />
M, Zhu F, Oluwadara O, Rao N, Braun J, Borneman J (2006)<br />
Abundant and diverse fungal microbiota in the murine intestine.<br />
Applied and Environmental Microbiology 72: 793–801.<br />
Seifert, KA, Lévesque CA (2004) Phylogeny and molecular diagnosis<br />
of mycotoxigenic fungi. European Journal of Plant Pathology<br />
110: 449–471.<br />
Seifert KA, Samson RA, deWaard JR, Houbraken J, Lévesque CA,<br />
Moncalvo J-M, Louis-Seize G and Hebert PDN (2007) Prospects<br />
for fungus identification using CO1 DNA barcodes, with<br />
Penicillium as a test case. Proceedings of the National Academy<br />
of Sciences, USA 104: 3901–3906.<br />
Shepard JR, Addison RM, Alexander BD, Della-Latta P, Gherna<br />
M, Haase G, Hall G, Johnson JK, Merz WG, Peltroche-<br />
Llacsahuanga H, Stender H, Venezia RA, Wilson D., Procop GW,<br />
Wu F, Fiandaca MJ (2008) Multicenter evaluation of the Candida<br />
albicans/Candida glabrata peptide nucleic acid fluorescent in situ<br />
hybridization method for simultaneous dual-color identification of<br />
C. albicans and C. glabrata directly from blood culture bottles.<br />
Journal of Clinical Microbiology 46: 50–55.<br />
Sholberg PL, O’Gorman D, Bedford K, Lévesque CA (2005) Development<br />
of a DNA macroarray for detection and monitoring of economically<br />
important apple diseases. Plant Disease 89: 1143–1150.<br />
188<br />
<br />
ima fUNGUS
Molecular identification and detection of fungi<br />
Sholberg PL, O’Gorman D, Bedford KE (2006) Monitoring Erwinia<br />
amylovora in pome fruit orchards using reverse dot-blot<br />
hybridization. Acta Horticulturae 704: 91-98.<br />
Stanley KK, Szewczuk E (2005) Multiplexed tandem PCR: gene<br />
profiling from small amounts of RNA using SYBR Green<br />
detection. Nucleic Acids Research 33: e180.<br />
Summerbell RC, Lévesque CA, Seifert KA, Bovers M, Fell JW, Diaz<br />
MR, Boekhout T, de Hoog GS, Stalpers J, Crous PW (2005)<br />
Microcoding: the second step in DNA barcoding. Philosophical<br />
Transactions of the Royal Scoiety, B 360: 1897–1903.<br />
Sun J, Li X, Zeng H, Xie Z, Lu C, Xi L, de Hoog GS (2010a)<br />
Development and evaluation of loop-mediated isothermal<br />
amplification (LAMP) for the rapid diagnosis of Penicillium<br />
marneffei in archived t<strong>issue</strong> samples. FEMS Immunology and<br />
Medical Microbiology 58: 381–388.<br />
Sun J, Najafzadeh MJ, Vicente V, Xi L, de Hoog GS (2010b)<br />
Rapid detection of pathogenic fungi using loop-mediated<br />
isothermal amplification, exemplified by Fonsecaea agents of<br />
chromoblastomycosis. Journal of Microbiological Methods 80:<br />
19–24.<br />
Szemes M, Bonants P, de Weerdt M, Baner J, Landegren U, Schoen<br />
CD (2005) Diagnostic application of padlock probes – multiplex<br />
detection of plant pathogens using universal microarrays. Nucleic<br />
Acids Research 33: e70.<br />
Tambong JT, de Cock AWAM, Tinker NA, Lévesque CA (2006)<br />
Oligonucleotide array for identification and detection of Pythium<br />
species. Applied and Environmental Microbiology 72: 2691–<br />
2706.<br />
Tatibana BT, Sano A, Uno J, Kamei K, Igarashi T, Mikami Y, Miyaji<br />
M, Nishimura K, Itano EN (2009) Detection of Paracoccidioides<br />
brasiliensis gp43 gene in sputa by loop-mediated isothermal<br />
amplification method. Journal of Clinical Laboratory Analysis 23:<br />
139–143.<br />
Tomita N, Mori Y, Kanda H, Notomi T (2008) Loop-mediated<br />
isothermal amplification (LAMP) of gene sequences and simple<br />
visual detection of products. Nature Protocols 3: 877–882.<br />
Tsui CKM, Wang B, Khadempour L, Alamouti SM, Bohlmann<br />
J, Murray BW, Hamelin RC (2010) Rapid identification and<br />
detection of pine pathogenic fungi associated with mountain pine<br />
beetles by padlock probes. Journal of Microbiological Methods<br />
83: 26–33.<br />
Tsui CKM, Wang B, Schoen C, Hamelin RC (2012) Padlock probe<br />
for the detection of pathogenic fungi. In Laboratory Protocols<br />
in Fungal Biology: Current Methods in Fungal Biology (Gupta<br />
VA, Tuohy M, Ayyachamy M, Turner KM, O’Donovan A, eds): in<br />
press. Springer-Verlag.<br />
Uehara T, Kushida A, Momota Y (1999) Rapid and sensitive<br />
identification of Pratylenchus spp. using reverse dot blot<br />
hybridization. Nematology 1: 549–555.<br />
Urakawa H, Fantroussi SE, Smidt H, Smoot JC, Tribou EH, Kelly<br />
JJ, Noble PA, Stahl DA (2003) Optimization of single-base-pair<br />
mismatch discrimination in oligonucleotide microarrays. Applied<br />
and Environmntal Microbiology 69: 2848–2856.<br />
van Doorn R, Szemes M, Bonants P, Kowalchuk GA, Salles JF,<br />
Ortenberg E, Schoen CD (2007) Quantitative multiplex detection<br />
of plant pathogens using a novel ligation probe-based system<br />
coupled with universal, high-throughput real-time PCR on<br />
OpenArrays ® . BMC Genomics 8: 276.<br />
van Doorn R, Slawiak M, Szemes M, Dullemans AM, Bonants<br />
P, Kowalchuk GA, Schoen CD (2009) Robust detection and<br />
identification of multiple oomycetes and fungi in environmental<br />
samples by using a novel cleavable padlock probe-based ligation<br />
detection assay. Applied and Environmental Microbiology 75:<br />
4185–4193.<br />
Wang B, Dwyer DE, Chew CB, Kol C, He ZP, Joshi H, Steain MC,<br />
Cunningham AL, Saksena NK (2009) Sensitive detection of the<br />
K103N non-nucleoside reverse transcriptase inhibitor resistance<br />
mutation in treatment-naive HIV-1 infected individuals by rolling<br />
circle amplification. Journal of Virological Methods 161: 128–135.<br />
Wang B, Dwyer DE, Blyth CC, Soedjono M, Shi H, Kesson A,<br />
Ratnamohan M, McPhie K, Cunningham AL, Saksena NK (2010)<br />
Detection of the rapid emergence of the H275Y mutation associated<br />
with oseltamivir resistance in severe pandemic influenza virus A/<br />
H1N1 09 infections. Antiviral Research 87: 16–21.<br />
Wang Y, Kang Z, Gao H, Gao Y, Qin L, Lin H, Yu F, Qi X, Wang X<br />
(2011) A one-step reverse transcription loop-mediated isothermal<br />
amplification for detection and discrimination of infectious bursal<br />
disease virus. Virology Journal 8: 108.<br />
Weber SD, Ludwig W, Schleifer K-H, Fried J (2007) Microbial<br />
composition and structure of aerobic granular sewage biofilms.<br />
Applied and Environmental Microbiology 73: 6233–6240.<br />
Wilson DA, Joyce MJ, Hall LS, Reller LB, Roberts GD, Hall GS,<br />
Alexander BD, Procop W (2005) Multicenter evaluation of<br />
a Candida albicans peptide nucleic acid fluorescent in situ<br />
hybridization probe for characterization of yeast isolates from<br />
blood cultures. Journal of Clinical Microbiology 43: 2909–2912.<br />
Xiong L, Kong F, Yang Y, Cheng J, Gilbert GL (2006) Use of PCR and<br />
reverse line blot hybridization macroarray based on 16S-23S<br />
rRNA gene internal transcribed spacer sequences for rapid<br />
identification of 34 Mycobacterium species. Journal of Clinical<br />
Microbiology 44: 3544–3550.<br />
Yang YP, Corley N, Garcia-Heras J (2001) Reverse dot-blot<br />
hybridization as an improved tool for the molecular diagnosis<br />
of point mutations in congenital adrenal hyperplasia caused by<br />
21-hydroxylase deficiency. Molecular Diagnostics 6: 193–199.<br />
Zhang N, Geiser MD, Smart CD (2007) Macroarray detection of<br />
solanaceous plant pathogens in the Fusarium solani species<br />
complex. Plant disease 91: 1612–1620.<br />
Zhang N, McCarthy ML, Smart CD (2008) A macroarray system for<br />
the detection of fungal and oomycete pathogens of solanaceous<br />
crops. Plant disease 92: 953–960.<br />
Zhang Y, Coyne MY, Will SG, Levenson CH, Kawasaki ES (1991)<br />
Single-base mutational analysis of cancer and genetic diseases<br />
using membrane bound modified oligonucleotides. Nucleic Acids<br />
Research 19: 3929–3933.<br />
Zhao K, Shi W, Han F, Xu Y, Zhu L, Zou Y, Wu X, Zhu H, Tan F,<br />
Tao S, Tang X (2011). Specific, simple and rapid detection of<br />
porcine circovirus type 2 using the loop-mediated isothermal<br />
amplification method. Virology Journal 8: 126.<br />
Zhou X, Kong F, Sorrell TC, Wang H, Duan Y, Chen SC (2008)<br />
Practical method for detection and identification of Candida,<br />
Aspergillus, and Scedosporium spp. by use of rolling-circle<br />
amplification. Journal of Clinical Microbiology 46: 2423–2427.<br />
ARTICLE<br />
volume 2 · no. 2<br />
189
ARTICLE<br />
<br />
ima fUNGUS
doi:10.5598/imafungus.2011.02.02.10 <strong>IMA</strong> <strong>Fungus</strong> · volume 2 · no 2: 191–199<br />
Advances in Glomeromycota taxonomy and classification<br />
Fritz Oehl 1 , Ewald Sieverding 2 , Javier Palenzuela 3 , Kurt Ineichen 4 , and Gladstone Alves da Silva 5<br />
1<br />
Agroscope Reckenholz-Tänikon Research Station (ART), Ecological Farming Systems, Reckenholzstrasse 191, CH-8046 Zürich, Switzerland;<br />
corresponding author e-mail: fritz.oehl@art.admin.ch<br />
2<br />
Institute of Plant Production and Agroecology in the Tropics and Subtropics, University of Hohenheim, Garbenstrasse 13, D-70593 Stuttgart,<br />
Germany<br />
3<br />
Departamento de Microbiología del Suelo y Sistemas Simbióticos, Estación Experimental del Zaidín, CSIC, Profesor Albareda 1, 18008<br />
Granada, Spain<br />
4<br />
Basel-Zürich Plant Science Center, Institute of Botany, University of Basel, Hebelstrasse 1, CH-4056 Basel, Switzerland<br />
5<br />
Departamento de Micologia, CCB, Universidade Federal de Pernambuco, Av. Prof. Nelson Chaves s/n, Cidade Universitaria, 50670-420,<br />
Recife, PE, Brazil<br />
ARTICLE<br />
Abstract: Concomitant morphological and molecular analyses have led to major breakthroughs in the taxonomic<br />
organization of the phylum Glomeromycota. Fungi in this phylum are known to form arbuscular mycorrhiza, and so<br />
far three classes, five orders, 14 families and 29 genera have been described. Sensu lato, spore formation in 10 of<br />
the arbuscular mycorrhiza-forming genera is exclusively glomoid, one is gigasporoid, seven are scutellosporoid,<br />
four are entrophosporoid, two are acaulosporoid, and one is pacisporoid. Spore bimorphism is found in three<br />
genera, and one genus is associated with cyanobacteria. Here we present the current classification developed in<br />
several recent publications and provide a summary to facilitate the identification of taxa from genus to class level.<br />
Key words:<br />
Archaeosporomycetes<br />
endomycorrhizas<br />
evolution<br />
Gigasporales<br />
Glomerales<br />
Glomeromycetes<br />
Paraglomeromycetes<br />
phylogeny<br />
VA mycorrhiza<br />
Article info: Submitted 10 November 2011; Accepted: 16 November 2011; Published: 18 November 2011.<br />
Introduction<br />
Glomeromycota taxonomy was largely morphologically driven<br />
up to the end of the last millennium. All glomeromycotean<br />
fungi, except one genus, are known to form arbuscular<br />
mycorrhiza. Their identification was based on spore<br />
morphology, spore formation, and spore wall structure<br />
(e.g. Gerdemann & Trappe 1974, Walker & Sanders 1986,<br />
Morton & Benny 1990, Schenck & Pérez 1990). However,<br />
as soon as molecular phylogenetic tools became available,<br />
they were included in taxonomic analyses (e.g. Simon et al.<br />
1992) and soon became the drivers of the establishment<br />
of a new taxonomy (Morton & Redecker 2001, Schüßler et<br />
al. 2001). In 1990, without the benefit of molecular aspects,<br />
the arbuscular mycorrhiza-forming fungi were organized<br />
in three families (Acaulosporaceae, Gigasporaceae, and<br />
Glomeraceae) and six genera (Acaulospora, Entrophospora,<br />
Gigaspora, Glomus, Sclerocystis, and Scutellospora)<br />
within one order, Glomerales (Morton & Benny 1990) of the<br />
fungal phylum Zygomycota. That classification was based<br />
on spore morphology and spore formation characteristics<br />
(acaulosporoid, entrophosporoid, gigasporoid, glomoid,<br />
radial-glomoid, and scutellosporoid). Differences in spore<br />
wall structure were used at the species level.<br />
Today, we accept three classes (Archaeosporomycetes,<br />
Glomeromycetes, and Paraglomeromycetes), five orders<br />
(Archaeosporales, Diversisporales, Gigasporales, Glomerales<br />
and Paraglomerales), 14 families, 29 genera and approximately<br />
230 species (e.g. Morton & Redecker 2001, Schüßler et al.<br />
2001, Oehl & Sieverding 2004, Walker & Schüßler 2004,<br />
Sieverding & Oehl 2006, Spain et al. 2006, Oehl et al. 2008,<br />
2011a–d, Palenzuela et al. 2008).<br />
Until recently, it was unclear whether glomoid and<br />
gigasporoid species could be further divided into different<br />
morphological groups congruent with the major phylogenetic<br />
clades obtained by molecular analyses. A first revision of the<br />
sporogenous cell forming (gigasporoid and scutellosporoid)<br />
Glomeromycetes according to concomitant morphological<br />
and phylogenetic features (Oehl et al. 2008) was not accepted<br />
by all mycologists (Morton & Msiska 2010). However, later<br />
studies with a broader database (e.g. Goto et al. 2010, 2011,<br />
Oehl et al. 2010, 2011b) confirmed that the revised genus<br />
Scutellospora, as well as the new Racocetra, Cetraspora,<br />
Dentiscutata, and Orbispora, are monophyletic.<br />
© 2011 International Mycological Association<br />
You are free to share - to copy, distribute and transmit the work, under the following conditions:<br />
Attribution:<br />
You must attribute the work in the manner specified by the author or licensor (but not in any way that suggests that they endorse you or your use of the work).<br />
Non-commercial: You may not use this work for commercial purposes.<br />
No derivative works: You may not alter, transform, or build upon this work.<br />
For any reuse or distribution, you must make clear to others the license terms of this work, which can be found at http://creativecommons.org/licenses/by-nc-nd/3.0/legalcode. Any of the above conditions can be waived if you get<br />
permission from the copyright holder. Nothing in this license impairs or restricts the author’s moral rights.<br />
volume 2 · no. 2 191
Oehl et al.<br />
ARTICLE<br />
A large group of species forms glomoid spores, and it<br />
had been believed that there were too few morphological<br />
characters of significance to differentiate them. Taxonomists<br />
have consequently started basing groupings of the glomoid<br />
species almost exclusively on molecular phylogenetic<br />
characters. A recent revision of these glomoid species<br />
has, however, shown that molecular phylogeny is actually<br />
congruent with the morphological characteristics of these<br />
fungi (Oehl et al. 2011c). Fungal species with entrophosporoid<br />
spore formation were also revised (Oehl et al. 2011d).<br />
The objective of this paper is to present the current overall<br />
classification system of Glomeromycota that has emerged<br />
from these recent studies, and to summarize the major<br />
morphological features in the phylum down to genus level.<br />
Materials and Methods<br />
The morphological, molecular, and phylogenetic analyses<br />
performed are presented in a series of recent publications<br />
dealing with different species groups of Glomeromycota (e.g.<br />
Oehl et al. 2006, 2010, 2011a, b, d, f, Sieverding & Oehl<br />
2006, Silva et al. 2006, Spain et al. 2006, Palenzuela et al.<br />
2008, 2010, 2011).<br />
Results and Discussion<br />
Figure 1 is a schematic tree for Glomeromycota based on<br />
molecular phylogenetic analyses of the SSU, ITS region, partial<br />
LSU of the rRNA gene, and partial β-tubulin gene (e.g. Oehl et<br />
al. 2008, 2010, 2011a–d). In Table 1, the major morphological<br />
features of all higher level taxa are presented, with the taxa<br />
arranged according to their taxonomic rank down to genus.<br />
Three glomeromycotean classes, five orders, 14 families, and<br />
29 genera have been recognized to date (Table 1). Sensu lato,<br />
spore formation in 10 of the arbuscular mycorrhiza-forming<br />
genera have exclusively glomoid, one has gigasporoid, seven<br />
have scutellosporoid, four have entrophosporoid, two genera<br />
have acaulosporoid, and one has pacisporoid spore formation,<br />
while three genera show spore bimorphism, and one genus<br />
is associated with cyanobacteria (the only one not forming<br />
arbuscular mycorrhizas).<br />
Hitherto, Paraglomeromycetes are monogeneric (Table<br />
1), are characterized by mono-walled spores formed<br />
terminally on hyphae (i.e. glomoid spores sensu lato), and<br />
germinate directly through the spore wall. Their arbuscular<br />
mycorrhizal structures do not or only faintly stain in trypan<br />
blue. Archaeosporomycetes includes organisms that are<br />
exclusively bimorphic since they form either acaulosporoid or<br />
entrophosporoid spores simultaneously with glomoid spores,<br />
or are associated with cyanobacteria. The mycorrhizal<br />
structures of Archaeosporaceae are similar to those of<br />
Paraglomeraceae, while Ambisporaceae form vesiculararbuscular<br />
mycorrhizal structures staining pale blue in trypan<br />
blue. In contrast, mycorrhizal structures in Glomeromycetes<br />
stain blue to dark blue in trypan blue. In Glomeromycetes,<br />
Gigasporales species do not form intraradical vesicles but<br />
auxiliary cells in soils, which clearly distinguish them from<br />
Glomerales and Diversisporales.<br />
Gigasporales exhibit gigasporoid or scutellosporoid spore<br />
formation (Oehl et al. 2011b), i.e. spores formed terminally<br />
on sporogenous cells and with either germ warts on the<br />
inner surface of the mono-walled spore wall (gigasporoid;<br />
Gigasporaceae), or a discrete germination shield on the<br />
innermost (= ‘germinal wall’) of 2–4 walls (scutellosporoid).<br />
There are three families with scutellosporoid spore formation<br />
(sensu lato): Dentiscutataceae, Racocetraceae and<br />
Scutellosporaceae (Oehl et al. 2008). Scutellosporaceae form<br />
mono-lobed (Orbispora) or bi-lobed (Scutellospora), hyaline<br />
germination shields (Figs 2–4). Racocetraceae species form<br />
wavy-like, multiply lobed, hyaline germination shields and<br />
have either two (Racocetra) or three (Cetraspora) spore walls<br />
(Figs 5–8). Dentiscutataceae species form yellow-brown to<br />
brown germ shields that are bi-lobed (Fuscutata; Fig. 9) or<br />
with multiple compartments (Dentiscutata, triple-walled;<br />
Quatunica four-walled; Figs 10–11).<br />
In Archaeosporales and Diversisporales, four genera<br />
have spore formation laterally on the neck of terminal or<br />
intercalary sporiferous saccules (= acaulosporoid sensu<br />
lato; Table 1): Acaulospora, Otospora, and the bi-morphic<br />
Ambispora and Archaeospora. These genera can easily be<br />
separated on spore wall number and spore wall structure<br />
(Palenzuela et al. 2008). Triple-walled Acaulospora<br />
species have a characteristic granular, ‘beaded’ inner wall<br />
surface (Morton & Benny 1990), which is absent in acauloambisporoid<br />
spores of triple-walled Ambispora species<br />
(Spain et al. 2006, Palenzuela et al. 2011). The wall structure<br />
of the bi-walled Otospora is more complex than that of biwalled<br />
Archaeospora species (Palenzuela et al. 2008).<br />
In Archaeosporales, Diversisporales, and Glomerales,<br />
there are five genera with spore formation within the<br />
neck of terminal or intercalary sporiferous saccules (i.e.<br />
entrophosporoid sensu lato; Table 1): Entrophospora,<br />
Kuklospora, Sacculospora, Tricispora, and bimorphic<br />
Intraspora (Oehl et al. 2011d). Triple-walled Kuklospora has<br />
the characteristic granular, ‘beaded’ inner wall surface of<br />
Acaulosporaceae (Sieverding & Oehl 2006), which is absent<br />
in spores of triple-walled Sacculospora (Oehl et al. 2011d).<br />
The wall structure of bi-walled Entrophospora and Tricispora<br />
is more complex than that of bi-walled, bimorphic Intraspora<br />
species (Sieverding & Oehl 2006, Oehl et al. 2011d).<br />
Entrophospora and Tricispora can be distinguished through<br />
the two cicatrices (scars) and pore structures proximal and<br />
distal to the sporiferous saccule: the proximal pore is wide<br />
in Tricispora and closed by a septum, while it is narrow and<br />
closed by a plug in Entrophospora. The distal pore and scar<br />
is absent in Entrophospora from the structural layer, and<br />
formed only on the overlying, hyaline, evanescent layer,<br />
while, in light microscopy, the distal pore with a distal scar is<br />
obvious in Tricispora (Sieverding & Oehl 2006, Palenzuela et<br />
al. 2010, Oehl et al. 2011d).<br />
192 ima fUNGUS
Advances in Glomeromycota classification<br />
ARTICLE<br />
Fig. 1. Representative tree of the phylum Glomeromycota based on molecular (SSU, ITS region, partial LSU of the rRNA gene, and partial<br />
β-tubuline gene) and morphological analyses (spore wall structures, structures of the spore bases and subtending hyphae, germination, and<br />
germination shield structures). Adapted from (Oehl et al. 2008, 2011a–d). The drawings in the central columns show the spore formation types<br />
of the genera, and the typical germination shields for those genera which form persistent shields already during spore formation.<br />
volume 2 · no. 2<br />
193
Oehl et al.<br />
ARTICLE<br />
Table 1. Major morphological characters for higher level taxa of Glomeromycota from class to genus level.<br />
Class<br />
Order<br />
Family Genus Spore formation Number of<br />
spore walls<br />
Germination; specific<br />
germination structure<br />
Mycorrhizal structures;<br />
staining in Trypan blue<br />
Glomeromycetes Germ tube (gt) Vesicles, Arbuscles,<br />
Hyphae<br />
Glomerales Glomeraceae Glomus Glomoid (terminally on hyphae)<br />
Funneliformis Glomoid<br />
Funneliformoid sensu stricto<br />
Septoglomus Glomoid<br />
Septoglomoid sensu stricto<br />
1<br />
1<br />
1<br />
gt through hypha<br />
gt through hypha<br />
gt through hyphae<br />
V, A, H<br />
V, A, H<br />
V, A, H<br />
Simiglomus Glomoid<br />
Simiglomoid sensu stricto<br />
1<br />
gt through hypha?<br />
V, A, H<br />
Entrophosporaceae Claroideoglomus Glomoid sensu lato<br />
Claroideoglomoid sensu stricto<br />
1<br />
gt through hypha<br />
V, A, H<br />
Albahypha Glomoid<br />
Claroideoglomoid sensu lato<br />
1<br />
gt through hypha?<br />
V, A, H<br />
Viscospora Glomoid<br />
Claroideoglomoid sensu lato<br />
1<br />
gt through hypha<br />
V, A, H<br />
Entrophospora Entrophosporoid (in the neck of a saccule)<br />
gt through wall?<br />
2<br />
V, A, H<br />
Diversisporales Diversisporaceae Diversispora Glomoid<br />
Diversisporoid sensu stricto<br />
1<br />
gt through hypha<br />
V, A, H<br />
Redeckera Glomoid<br />
(Diversisporo-)Redeckeroid sensu stricto<br />
1<br />
gt through hypha?<br />
V, A, H<br />
Otospora Acaulosporoid (on the neck of sporiferous<br />
saccule): otosporoid sensu stricto 2<br />
Unknown?<br />
V, A, H<br />
Tricispora Entrophosporoid<br />
Tricisporoid sensu stricto 2<br />
Unknown?<br />
V, A, H<br />
Sacculosporaceae Sacculospora Entrophosporoid<br />
Sacculosporoid sensu stricto 3<br />
Unknown?<br />
V, A, H<br />
Pacisporaceae Pacispora Pacisporoid<br />
2<br />
gt through wall; multiply<br />
lobed germ structure V, A, H<br />
Acaulosporaceae Kuklospora Entrophosporoid<br />
Kuklosporoid sensu stricto 3<br />
Acaulospora Acaulosporoid<br />
3<br />
gt through wall; monolobed,<br />
hyaline germ shield<br />
(=orb)<br />
gt through wall; mono-(to<br />
multiply) lobed, hyaline<br />
germ shield (=orb)<br />
V, A, H<br />
V, A, H<br />
194 ima fUNGUS
Advances in Glomeromycota classification<br />
ARTICLE<br />
Gigasporales Scutellosporaceae Orbispora Scutellosporoid (on sporogenous cells, and<br />
forming germ shields); Orbisporoid sensu<br />
stricto<br />
Scutellospora Scutellosporoid<br />
Dentiscutataceae Fuscutata Scutellosporoid<br />
Fuscutatoid sensu stricto<br />
Dentiscutata Scutellosporoid<br />
Dentiscutatoid sensu stricto<br />
Quatunica Scutellosporoid<br />
Dentiscutatoid sensu stricto<br />
Racocetraceae Cetraspora Scutellosporoid<br />
Racocetroid sensu stricto<br />
Racocetra Scutellosporoid<br />
Racocetroid sensu stricto<br />
Gigasporaceae Gigaspora Gigasporoid (on sporogenous cells, and<br />
forming germ warts) 1<br />
Archaeosporomycetes<br />
Archaeosporales Ambisporaceae Ambispora Bimorph: Acaulo- & Glomo-ambisporoid<br />
Archaeosporaceae Archaeospora Bimorph: Acaulo- & Glomo-archaeosporoid<br />
Intraspora Bimorph: Entropho-& Glomo-intrasporoid<br />
Geosiphonaceae Geosiphon Glomoid sensu lato<br />
Paraglomeromycetes<br />
Paraglomerales Paraglomeraceae Paraglomus Glomoid sensu lato<br />
3<br />
3<br />
3<br />
3<br />
4<br />
3<br />
2<br />
3 (Ac)<br />
1 (Gl)<br />
2 (Ac)<br />
1 (Gl)<br />
2 (Ac)<br />
1 (Gl)<br />
1<br />
1<br />
gt through wall; monolobed,<br />
hyaline germ shield<br />
(=orb)<br />
gt through wall; bi-lobed,<br />
hyaline, violin-shaped germ<br />
shield<br />
A, H<br />
A, H<br />
gt through wall; bi-lobed,<br />
brown, oval shield A, H<br />
gt through wall; brown germ<br />
shield with multiple small<br />
compartments<br />
gt through wall; brown<br />
germ shield with multiple<br />
compartments<br />
A, H<br />
A, H<br />
gt through wall; multiply<br />
lobed, hyaline germ shield A, H<br />
gt through wall; multiply<br />
lobed, hyaline germ shield A, H<br />
gt through wall; germ warts<br />
on inner spore wall layer A, H<br />
Multiply-lobed germ<br />
structure (Ac) & gt through<br />
hypha (Gl)<br />
V, A, H<br />
gt through wall; germ trunk<br />
(Ac), & gt through hypha<br />
(Gl)?<br />
A, H<br />
Unknown?<br />
A, H<br />
gt through hypha? Associated with<br />
cyanobacteria<br />
gt through wall<br />
A, H<br />
volume 2 · no. 2<br />
195
Oehl et al.<br />
ARTICLE<br />
Figs 2–11. Characteristic germination shields in Gigasporales with germ pore (gp) as connection between spore cell contents and shields that<br />
are positioned on the surface of the germinal wall; germ tubes emerge from germ tube initiations (gti). Fig. 2. Orbispora pernambucana (isotype,<br />
ZT Myc 641) with mono-lobed, hyaline germ shield (orb). Figs 3–4. Scutellospora calospora (photo taken at INVAM) and S. dipurpurescens<br />
(holotype OSC #83343) have bi-lobed, violin-shaped, hyaline shields. Figs 5–8. Racocetra coralloidea (type, OSC #31026), R. castanea (ex<br />
type, ZT Myc 4377), Cetraspora nodosa (isotype, DPP, Szczecin, Poland) and C. helvetica (isotype, ZT Myc 3038) have wavy-like, multiply<br />
lobed, hyaline shields. Figs 9–11. Dentiscutataceae shields are yellow brown to brown. Fig. 9. Dentiscutata reticulata (photo taken at INVAM)<br />
shields with multiple small compartments. Fig. 10. Quatunica erythropa (photo taken at INVAM) is assumed to be the only known species in<br />
Glomeromycota with four spore walls. Fig. 11. Fuscutata heterogama (ex type, ZT Myc 642) has a bi-lobed, oval to ovoid shield.<br />
196 ima fUNGUS
Advances in Glomeromycota classification<br />
ARTICLE<br />
Figs 12–21. Characteristic spore bases and subtending hyphae (sh) in Glomeromycetes genera with glomoid spore formation. Figs 12–13.<br />
Glomus ambisporum (Oehl collection, from Bolivia) and G. aureum (type, ZT Myc 822) with two wall layers (SWL1 and SWL2), marked introverted<br />
wall thickening at sb and in sh, and a small, bridging septum (sp). Fig. 14. Funneliformis coronatus (ex type, Oehl collection) with funnelshaped<br />
sh and conspicuous sp; introverted wall thickening is lacking. Fig. 15. Septoglomus constrictum (Oehl collection, from Switzerland) with<br />
conspicuous septum that sometimes resembles a plug. Fig. 16. Simiglomus hoi (Oehl collection, specimen mounted at York university) with<br />
cylindrical sh; sh wall thickened over long distances; several septae are regularly observed within the hyphae; no introverted wall thickening<br />
at sb, pore at sb generally opened. Fig. 17. Claroideoglomus etunicatum (Oehl collection, from Bolivia) with funnel/bill-shaped, white sh; all<br />
Entrophosporaceae (syn. Claroideoglomeraceae) with characteristic color change of structural wall layer at sb, if spores are pigmented. Fig.<br />
18. Albahypha drummondii (type, DPP) with slightly funnel-shaped, white sh. Fig. 19. Viscospora viscosa (ex type, photo taken at INVAM) with<br />
cylindrical, white hypha; sp within sh in some distance of sb; introverted wall thickening of sh at sp position, here not that obvious as usually found<br />
for this species; viscose spore surface. Fig. 20. Diversispora versiformis with short, fragile sh that is principally continuous with semi-persistent<br />
outermost spore wall layer (SWL1) but not with structural layer SWL2 (Oehl collection, from Tibet). Fig. 21. Redeckera fulva (Oehl ex Trappe<br />
collection) with inflating sh and conspicuous broad sp exactly at spore base.<br />
In Diversisporales and Glomerales, 10 genera exclusively<br />
differentiate mono-walled, glomoid (9) or bi-walled pacisporoid<br />
(1) spores, all formed on subtending hyphae (Oehl & Sieverding<br />
2004, Oehl et al. 2011a). The morphological differentiation of<br />
the glomoid species is mainly based on the morphology of the<br />
subtending hyphae of the spores, and spore wall structure.<br />
Spores of Funneliformis, Glomus, Septoglomus, and Simiglomus<br />
species have subtending hyphae that are concolorous or slightly<br />
lighter in colour than the spore wall (Table 1, Figs 12–16).<br />
Albahypha, Claroideoglomus, and Viscospora form spores in<br />
which the structural wall layer is continuous with the subtending<br />
hyphal wall layer, but the subtending hyphae are hyaline (Figs<br />
17–19). In contrast, Diversispora and Redeckera form spores<br />
whose structural wall layer is not obviously continuous with<br />
the hyphal wall layer (Figs 20–21); consequently, such spores<br />
appear to have included ‘endospores’.<br />
volume 2 · no. 2<br />
197
Oehl et al.<br />
ARTICLE<br />
Funneliformis, Glomus, Septoglomus, and Simiglomus<br />
can be separated by the structure of the spore base and<br />
subtending hyphae (sh). Glomus species often have an<br />
introverted wall thickening (Oehl et al. 2011a; Figs 12–13)<br />
which is only otherwise seen in Viscospora. Funneliformis<br />
species generally have an easily visible septum in the area<br />
of the spore base, and their sh are regularly funnel-shaped to<br />
cylindrical (Fig. 14). Septoglomus species have constricted<br />
to cylindrical sh, and usually there is a septum at the spore<br />
base (Fig. 15). In Simiglomus, sh are cylindrical and thickwalled,<br />
and they have several septa some distance from the<br />
spore base (Fig. 16). Claroideoglomus has funnel- to birdbill-shaped<br />
sh, with sh and sh walls that are > 2.5 times wider<br />
at the spore base than some distance from the base (Fig.<br />
17). Albahypha has slightly funnel to bill-shaped sh and sh<br />
walls that are < 2.0 times wider at the spore base than at<br />
some distance from the base (Fig. 18), and Viscospora has<br />
cylindrical sh (Fig. 19) with an sh wall that may be thickened<br />
over large distances and may bear septa in the hyphae<br />
with introverted wall thickenings in the area of the septum.<br />
In Diversispora, the sh are usually quite fragile and hyaline,<br />
distal to the pore closure at the spore base or in the sh (Fig.<br />
20). Redeckera species have a broad septum at the spore<br />
base (Fig. 21), and the structural wall layer does not continue<br />
more than 5–15 µm into the subtending hypha, and thus, the<br />
sh may inflate at this distance from the spore base.<br />
There are three bi-morphic genera with glomoid spore<br />
formation. Glomo-ambisporoid spores have a subhyaline<br />
to ochraceous, evanescent outer wall layer continuous with<br />
the outer acaulo-ambisporoid spore wall, while the second,<br />
structural layer is hyaline and continuous with the middle wall<br />
of acaulo-ambisporoid spores (Spain et al. 2006, Palenzuela<br />
et al. 2011). Glomo-archaeosporoid and Glomo-intrasporoid<br />
spores are among the smallest within Glomeromycota (ca. 30<br />
µm), and thus difficult to observe.<br />
FURB, Santa Catarina State, Brazil), GINCO-BEL in Louvain-<br />
La-Neuve (Glomeromycota In Vitro Collection at the Catholic<br />
University of Louvain, Belgium), or SAF in Zurich (Swiss<br />
Collection of Arbsucular Mycorrhizal Fungi at Agroscope<br />
ART, Switzerland) will facilitate further progresses in the<br />
taxonomy of glomeromycotean fungi that were thought to<br />
have not enough criteria to morphologically separate them<br />
unequivocally into the higher level taxa they phylogenetically<br />
belong to. Currently, several arbuscular mycorrhizal fungi<br />
are being described as new to science each year by an<br />
increasing numbers of research groups. A simple, but well<br />
justified conclusion is that, as a result of future concomitant<br />
morphological and molecular analyses, yet more higher level<br />
taxa will be proposed in this ancient fungal phylum, at all<br />
levels from class down to genus.<br />
AcknowledgEments<br />
We acknowledge the help of many current and former students and<br />
technicians in Switzerland, Spain, and Brazil for outstanding support<br />
(especially David Schneider, Robert Bösch, Giacomo Busco, Louis<br />
Lawouin, Domingo Alvarez, Danielle Silva, Natália Sousa, and<br />
Daniele Magna). This study has been supported within the Swiss<br />
National Science Foundation (SNSF) Project 315230_130764/1,<br />
by the SNSF Programme NFP48 ‘Landscapes and habitats of the<br />
Alps’, by the Spanish Ministry of Environment (MMA-OAPN, project<br />
70/2005) and FAECA (Junta de Andalucía, Spain, Project 92162/11),<br />
and by the Fundação de Amparo à Ciência e Tecnologia do Estado<br />
de Pernambuco (FACEPE) and the UFPE which provided grants<br />
to F. Oehl as ‘visiting professor’, and FACEPE which also provided<br />
financial support for G.A. Silva.<br />
REFERENCES<br />
Perspectives<br />
Further separations of genera and families can be expected<br />
in the near future since many species and several species<br />
groups have not yet been analyzed by molecular phylogenetic<br />
methods (e.g. Glomus group Ab1, sensu Oehl et al. 2011a).<br />
Major efforts are needed to properly describe the morphology<br />
of, in particular, small-spored Glomus species (Błaszkowski<br />
et al. 2009a, b, 2010a, b), and it is difficult to predict how<br />
morphological identification will develop in those fungi.<br />
Other recent progress has been made on Acaulospora<br />
species with pitted surface ornamentation, where several<br />
species, that superficially all resembled A. scrobiculata,<br />
have been separated through extensive morphological and<br />
molecular spore analyses (e.g. Oehl et al. 2006, 2011e, f).<br />
The establishment of international and national collections of<br />
arbuscular mycorrhizal fungi, such as INVAM in Morgantown<br />
(International Culture Collection of (Vesicular) Arbuscular<br />
Mycorrhizal Fungi, West Virginia State University, USA), CICG<br />
in Blumenau (International Collection of Glomeromycota at<br />
Błaszkowski J, Kovács GM, Balázs TK (2009a) Glomus perpusillum,<br />
a new arbuscular mycorrhizal fungus. Mycologia 101: 247–255.<br />
Błaszkowski J, Ryszka P, Oehl F, Koegel S, Wiemken A, et al. (2009b)<br />
Glomus achrum and G. bistratum, two new species of arbuscular<br />
mycorrhizal fungi (Glomeromycota). Botany 87: 260–271.<br />
Błaszkowski J, Wubet T, Harikumar VS, Ryszka P, Buscot F (2010a)<br />
Glomus indicum, a new arbuscular mycorrhizal fungus. Botany<br />
88: 132–143.<br />
Błaszkowski J, Kovács GM, Balázs TK, Orlowska E, Sadravi M, et al.<br />
(2010b) Glomus africanum and G. iranicum, two new species of<br />
arbuscular mycorrhizal fungi (Glomeromycota). Mycologia 102:<br />
1450–1462.<br />
Gerdemann JW, Trappe JM (1974) The Endogonaceae of the Pacific<br />
Northwest. Mycologia Memoir 5: 1–76.<br />
Goto BT, Silva GA, Maia LC, Oehl F (2010) Dentiscutata colliculosa,<br />
a new species in the Glomeromycetes from Northeastern Brazil<br />
with colliculate spore ornamentation. Nova Hedwigia 90: 383–<br />
393.<br />
Goto BT, Silva GA, Maia LC, Souza RG, Coyne D, et al. (2011)<br />
Racocetra tropicana, a new species in the Glomeromycetes from<br />
tropical areas. Nova Hedwigia 92: 69–82.<br />
198 ima fUNGUS
Advances in Glomeromycota classification<br />
Morton JB, Benny GL (1990) Revised classification of arbuscular<br />
mycorrhizal fungi (Zygomycetes): a new order, Glomales, two<br />
new suborders, Glomineae and Gigasporineae, and two families,<br />
Acaulosporaceae and Gigasporaceae, with an emendation of<br />
Glomaceae. Mycotaxon 37: 471–491.<br />
Morton JB, Redecker D (2001) Two new families of Glomales,<br />
Archaeosporaceae and Paraglomaceae, with two new genera<br />
Archaeospora and Paraglomus, based on concordant molecular<br />
and morphological characters. Mycologia 93: 181–195.<br />
Morton JB, Msiska Z (2010) Phylogenies from genetic and<br />
morphological characters do not support a revision of<br />
Gigasporaceae (Glomeromycota) into four families and five<br />
genera. Mycorrhiza 20: 483–496.<br />
Oehl F, Sieverding E (2004) Pacispora, a new vesicular-arbuscular<br />
mycorrhizal fungal genus in the Glomeromycetes. Journal of<br />
Applied Botany and Food Quality 78: 72–82.<br />
Oehl F, Sýkorová Z, Redecker D, Wiemken A, Sieverding E (2006)<br />
Acaulospora alpina, a new arbuscular mycorrhizal fungal species<br />
characteristic for high mountainous and alpine regions of the<br />
Swiss Alps. Mycologia 98: 286–294.<br />
Oehl F, Souza FA, Sieverding E (2008) Revision of Scutellospora<br />
and description of five new genera and three new families in the<br />
arbuscular mycorrhiza-forming Glomeromycetes. Mycotaxon<br />
106: 311–360.<br />
Oehl F, Jansa J, Souza FA, Silva GA (2010) Cetraspora helvetica,<br />
a new ornamented species in the Glomeromycetes from Swiss<br />
agricultural fields. Mycotaxon 114: 71–84.<br />
Oehl F, Silva GA, Goto BT, Sieverding E (2011a) Glomeromycetes:<br />
three new genera and glomoid species reorganized. Mycotaxon<br />
116: 75–120.<br />
Oehl F, Silva DKA, Maia LC, Sousa NMF, Vieira HEE, Silva GA<br />
(2011b) Orbispora gen. nov., ancestral in the Scutellosporaceae<br />
(Glomeromycetes). Mycotaxon 116: 161–169.<br />
Oehl F, Silva GA, Goto BT, Maia LC, Sieverding E (2011c)<br />
Glomeromycota: two new classes and a new order. Mycotaxon<br />
116: 365–379.<br />
Oehl F, Silva GA, Sánchez-Castro I, Goto BT, Maia LC, Vieira<br />
HEE, Barea JM, Sieverding E, Palenzuela J (2011d) Revision<br />
of Glomeromycetes with entrophosporoid and glomoid spore<br />
formation with three new genera. Mycotaxon 117: 297–316.<br />
Oehl F, Sýkorová Z, Błaszkowski J, Sánchez-Castro I, Coyne D, et<br />
al. (2011e) Acaulospora sieverdingii, an ecologically diverse new<br />
fungus in the Glomeromycota, described from lowland temperate<br />
Europe and tropical West Africa. Journal of Applied Botany and<br />
Food Quality 84: 47–53.<br />
Oehl F, Silva GA, Palenzuela J, Sánchez-Castro I, Castillo C,<br />
Sieverding E (2011f) Acaulospora punctata, a new fungal species<br />
in the Glomeromycetes from mountainous altitudes of the Swiss<br />
Alps and Chilean Andes. Nova Hedwigia 93: 353–362.<br />
Palenzuela J, Ferrol N, Boller T, Azcón-Aguilar C, Oehl F (2008)<br />
Otospora bareai, a new fungal species in the Glomeromycetes<br />
from a dolomitic shrub-land in the National Park of Sierra de<br />
Baza (Granada, Spain). Mycologia 100: 282–291.<br />
Palenzuela J, Barea JM, Ferrol N, Azcón-Aguilar C, Oehl F (2010)<br />
Entrophospora nevadensis, a new arbuscular mycorrhizal<br />
fungus, from Sierra Nevada National Park (southeastern Spain).<br />
Mycologia 102: 624–632.<br />
Palenzuela J, Barea JM, Ferrol N, Oehl F (2011) Ambispora<br />
granatensis, a new arbuscular mycorrhizal fungus, associated<br />
with Asparagus officinalis in Andalucía (Spain). Mycologia 103:<br />
333–340.<br />
Schenck NC, Pérez Y (1990) Manual for the Identification of<br />
VA Mycorrhizal Fungi. 3rd edn: Gainesville, FL: Synergistic<br />
Publications.<br />
Schüßler A, Schwarzott D, Walker C (2001) A new fungal phylum,<br />
the Glomeromycota: phylogeny and evolution. Mycological<br />
Research 105: 1413–1421.<br />
Sieverding E, Oehl F (2006) Revision of Entrophospora and<br />
description of Kuklospora and Intraspora, two new genera in<br />
the arbuscular mycorrhizal Glomeromycetes. Journal of Applied<br />
Botany and Food Quality 80: 69–81.<br />
Silva GA, Lumini E, Maia LC, Bonfante P, Bianciotto V (2006)<br />
Phylogenetic analysis of Glomeromycota by partial LSU rDNA<br />
sequences. Mycorrhiza 16: 183–189.<br />
Simon L, Lalonde M, Bruns TD (1992) Specific amplification<br />
of 18S fungal ribosomal genes from vesicular-arbuscular<br />
endomycorrhizal fungi colonizing roots. Applied and<br />
Environmental Microbiology 58: 291–295.<br />
Spain JL, Sieverding E, Oehl F (2006) Appendicispora, a new genus<br />
in the arbuscular mycorrhizal-forming Glomeromycetes, with a<br />
discussion of the genus Archaeospora. Mycotaxon 97: 163–182.<br />
Walker C, Sanders FE (1986) Taxonomic concepts in the<br />
Endogonaceae: III. The separation of Scutellospora gen. nov.<br />
from Gigaspora Gerd. and Trappe. Mycotaxon 27: 169–182.<br />
Walker C, Schüssler A (2004) Nomenclatural clarifications and new<br />
taxa in the Glomeromycota. Mycological Research 108: 979–<br />
982.<br />
ARTICLE<br />
volume 2 · no. 2<br />
199
All submitted materials must be digitized and submitted electronically to d.hawksworth@nhm.ac.uk and p.crous@cbs.knaw.nl, with the<br />
manuscript ideally in Microsoft Word. Illustrations (Line drawings (600 dpi or higher) and half tone pictures (300 dpi or higher) should be<br />
submitted separately, never embedded in Word files. Phylogenetic trees will only be accepted when submitted as Powerpoint files, or in Adobe<br />
(never as pictures).<br />
The corresponding author should confirm that: (a) all named authors have agreed to publication of the work; and (b) the manuscript does not<br />
infringe any personal or other copyright or property rights.<br />
Papers cited as “in press” should be provided for the benefit of the referees.<br />
The content should be structured as follows: ABSTRACT, INTRODUCTION, MATERIALS AND METHODS, RESULTS, TAXON-<br />
OMY, DISCUSSION, ACKNOWLEDGEMENTS and REFERENCES. Key words and a Running Head should also be provided.<br />
English-English is preferred, and used for all non-article material. Authors of articles can use American-English, provided that it is consistent<br />
within their contributions. Words of non-English origin, like bona fide, prima facie, in vitro, in situ, should be placed in italic, together with<br />
scientific names of any rank (e.g. Ascomycota, Dothideales, Mycosphaerellaceae, Mycosphaerella nubilosa).<br />
Common abbreviations are as follows: h, min, s, mL, µL, mg/L, °C, Fig., d, wk, but also ITS, RPD, RFLP, rDNA, 18S etc.<br />
Authorities of fungal taxa should be omitted from the general text, unless novelties and synonymies are listed, or nomenclatural <strong>issue</strong>s<br />
discussed. In these cases, authorities for taxa should follow the list of authors’ names, see http://www.speciesfungorum.org/AuthorsOf-<br />
FungalNames.htm, followed by the year of publication of the name. Journal abbreviations in the text (species synonymies, descriptions,<br />
etc.) should follow the International Plant Name Index (see http://www.ipni.org/index.html).<br />
Experimental procedures must be reproducible and must follow Good Cultural Practice (Mycological Research 106: 1378–1379), with<br />
sequences lodged at GenBank, alignments in TreeBASE, voucher specimens in a public reference collection recognized in Index, or another<br />
recognized in Index Herbariorum (online) or the World Directory of Collections and Cultures of Microorganisms (online), with acronym and accession<br />
numbers (where allocated), and ex-type and other representative cultures in long-term genetic resource depositaries or collections. For<br />
new scientific names a MycoBank number is required (see www.MycoBank.org), and should be obtained on acceptance for publication.<br />
Collections must be cited as:<br />
Specimens examined. Country: State/Province/County: location, substratum, date (e.g. 10 Dec. 1993), collector (e.g. S. Uper & F. Ungus)<br />
(COLLECTION ACRONYM and ACCESSION NUMBER -- holotypus; cultures ex-holotype COLLECTION ACRONYM(S) and<br />
ACCESSION NUMBER(S)).<br />
Reference citation in text: References in the text should be chronological, and given in the following form: “Smith & Jones (1965) have<br />
shown ...”, or, “some authors (Zabetta 1928, Taylor & Palmer 1970, Zabetta 1970) consider that ...”. The names of collaborating authors are<br />
joined by an ampersand (&). Where there are three or more authors, names should be cited by the first name only, adding “et al.”, e.g. “Bowie,<br />
Black & White (1964)” are given as “Bowie et al. (1964)” or “(Bowie et al. 1964)”. Where authors have published more than one work in a<br />
year, to which reference is made, they should be distinguished by placing a, b, etc. immediately after the date, e.g. “Dylan (1965a, b)”. Reference<br />
citations in text should be in ascending order of year first, followed by authors’ names. Each reference should include the full title of the<br />
paper and journal, volume number, and the final as well as the first page number. In the case of chapters in books, the names of editors, first<br />
and last page numbers of the articles, publisher and place of publication are needed.<br />
Examples:<br />
Black JA, Taylor JE (1999a) Article title. Studies in Mycology 13: 1–10.<br />
Black JA, Taylor JE (In press) Article title. Fungal Biology.<br />
Black JA, Taylor JE, White DA (1981) Article title. In: Book Title (KA Seifert, W Jones & P. Jones, eds): 11–30. Town (and country if not<br />
well-known): Press.<br />
Simpson H, Seifert KA (2000) Book Title. 2nd edn. Town (and country if not well-known): Press.<br />
White DA (2001) Dissertation Title. PhD thesis, Department, University, Country.<br />
Copyright / Proprietary Rights Notice:<br />
Unless otherwise noted, the <strong>IMA</strong> holds the copyright on all materials published in <strong>IMA</strong> <strong>Fungus</strong>, whether in print or electronic form, both as<br />
a compilation and as individual articles. All Journal content is subject to ‘fair use’ provisions of U.S. or applicable international copyright laws<br />
(see http://www.copyright.gov/title17/92chap1.html). The following guidelines apply to individual users:<br />
A. Individuals may view, download, print, or save Journal content for the purposes of research, teaching, and/or private study. Systematic<br />
downloading (by robots or other automatic processes) is prohibited without explicit Publisher approval.<br />
B. Individuals may not reproduce, post, redistribute, sell, modify or create a derivative work of any Journal content, without prior, express<br />
written permission of the Publisher. Permission is granted, however, to provide a limited amount of print or electronic Journal content for<br />
purposes of regulatory approval, patent and/or trademark applications or other legal or regulatory purposes.<br />
C. Any use and/or copies of this Journal in whole or in part, must include the customary bibliographic citation, including author attribution,<br />
date, article title, the Journal name and its URL (web site address) and MUST include the copyright notice. Individuals may not<br />
remove, cover, overlay, obscure, block, or change any copyright notices, legends, or terms of use.<br />
D. For permissions to reprint or copy Journal content beyond that permitted by Section 107 or 108 of the U.S. Copyright Law, contact<br />
the Copyright Clearance Center. The fee code for users of the Transactional Reporting Service appears in each abstract and full text<br />
article.<br />
INSTRUCTIONS TO AUTHORS<br />
EDITORIAL BOARD
Editorial<br />
One <strong>Fungus</strong> One Name: a plant pathologists’s view (39)<br />
News<br />
1000 fungal genomes to be sequenced – The Ug99 stem rust (Puccinia graminis) of wheat threatens global supplies<br />
– A DNA barcode for Fungi proposed for adoption – New world record for the largest fungal basidiome – International<br />
meetings planned to discuss fungal nomenclature – MycoKeys<br />
Reports<br />
Genomics in China – International Association for Lichenology (IAL) – XVI Congress of European Mycologists<br />
(CEM XVI) – 30 th ECCO Annual Meeting in Utrecht – Mycological Society of America – 20 th Nordic Mycological<br />
Congress (NMC 20)<br />
(41)<br />
(46)<br />
Awards and Personalia<br />
<strong>IMA</strong> Young Mycologist Awards 2011 (Ethel Mary Doidge Medal; Keisuke Tubaki Medal; Daniel McAlpine Medal;<br />
Arthur Henry Reginald Buller Medal)<br />
(52)<br />
Distinguished Asian Mycologist Award: Kevin Hyde (53)<br />
Birthday Greeting: Jiang-Chun Wei’s 80 th (54)<br />
In Memoriam: Aino Marjatta Henssen (1925–2011); Zang Mu (1930–2011) (55)<br />
Research News<br />
Horizontal Gene Transfer (HGT) from Fungi is the basis for plant pathogenicity in oomycetes (57)<br />
Fungal pathogens as a driver of tree species diversity in tropical forests (58)<br />
A new system for the arbuscular mycorrhizal fungi (Glomeromycota) (59)<br />
New insights into global fungal species numbers? (59)<br />
Powdery mildews under scrutiny (60)<br />
Book News (62)<br />
Forthcoming Meetings (67)<br />
Articles<br />
“One <strong>Fungus</strong> = One Name: DNA and fungal nomenclature twenty years after PCR” by John W. Taylor 113<br />
“Penicillium menonorum, a new species related to P. pimiteouiense” by Stephen W. Peterson, Samantha S. Orchard, 121<br />
and Suresh Menon<br />
“What is Scirrhia?” by Pedro W. Crous, Andrew M. Minnis, Olinto L. Pereira, Acelino C. Alfenas, Rafael F. Alfenas, 127<br />
Amy Y. Rossman, and Johannes Z. Groenewald<br />
“A new species of Antherospora supports the systematic placement of its host plant” by Marcin Piątek, Matthias Lutz, 135<br />
Paul A. Smith, and Arthur O. Chater<br />
“Ascospore discharge, germination and culture of fungal partners of tropical lichens, including the use of a novel culture<br />
technique” by Ek Sangvichien, David L. Hawksworth, and Anthony J.S. Whalley<br />
143<br />
“A new dawn for the naming of fungi: impacts of decisions made in Melbourne in July 2011 on the future publication 155<br />
and regulation of fungal names” by David L. Hawksworth<br />
“The inclusion of downy mildews in a multi-locus-dataset and its reanalysis reveals a high degree of paraphyly in<br />
163<br />
Phytophthora” by Fabian Runge, Sabine Telle, Sebastian Ploch, Elizabeth Savory, Brad Day, Rahul Sharma, and<br />
Marco Thines<br />
“Validation and justification of the phylum name Cryptomycota phyl. nov.” by Meredith D.M. Jones, Thomas A. Richards,<br />
David L. Hawksworth, and David Bass<br />
173<br />
“Molecular techniques for pathogen identification and fungus detection in the environment” by Clement K.M. Tsui, 177<br />
James Woodhall, Wen Chen, C. André Lévesque, Anna Lau, Cor D. Schoen, Christiane Baschien, Mohammad<br />
Najafzadeh, and G. Sybren de Hoog<br />
“Advances in Glomeromycota taxonomy and classification” by Fritz Oehl, Ewald Sieverding, Javier Palenzuela, Kurt 191<br />
Ineichen, and Gladstone Alves da Silva