Modeling hemodynamic response for analysis of functional MRI time ...
Modeling hemodynamic response for analysis of functional MRI time ...
Modeling hemodynamic response for analysis of functional MRI time ...
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
Rajapakse et al. <br />
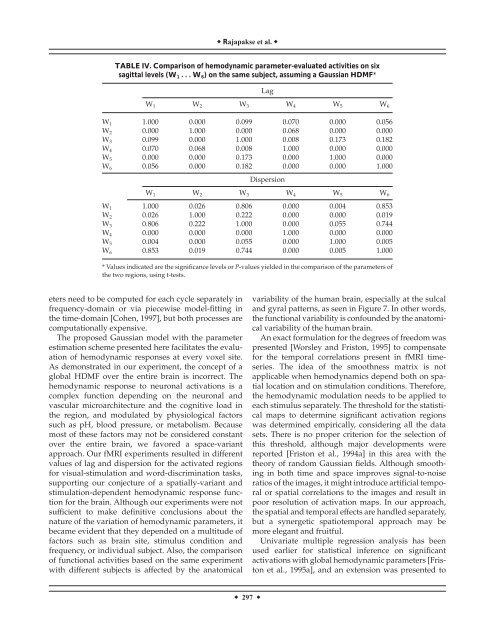
TABLE IV. Comparison <strong>of</strong> <strong>hemodynamic</strong> parameter-evaluated activities on six<br />
sagittal levels (W 1 ...W 6 ) on the same subject, assuming a Gaussian HDMF*<br />
Lag<br />
W 1 W 2 W 3 W 4 W 5 W 6<br />
W 1 1.000 0.000 0.099 0.070 0.000 0.056<br />
W 2 0.000 1.000 0.000 0.068 0.000 0.000<br />
W 3 0.099 0.000 1.000 0.008 0.173 0.182<br />
W 4 0.070 0.068 0.008 1.000 0.000 0.000<br />
W 5 0.000 0.000 0.173 0.000 1.000 0.000<br />
W 6 0.056 0.000 0.182 0.000 0.000 1.000<br />
Dispersion<br />
W 1 W 2 W 3 W 4 W 5 W 6<br />
W 1 1.000 0.026 0.806 0.000 0.004 0.853<br />
W 2 0.026 1.000 0.222 0.000 0.000 0.019<br />
W 3 0.806 0.222 1.000 0.000 0.055 0.744<br />
W 4 0.000 0.000 0.000 1.000 0.000 0.000<br />
W 5 0.004 0.000 0.055 0.000 1.000 0.005<br />
W 6 0.853 0.019 0.744 0.000 0.005 1.000<br />
* Values indicated are the significance levels or P-values yielded in the comparison <strong>of</strong> the parameters <strong>of</strong><br />
the two regions, using t-tests.<br />
eters need to be computed <strong>for</strong> each cycle separately in<br />
frequency-domain or via piecewise model-fitting in<br />
the <strong>time</strong>-domain [Cohen, 1997], but both processes are<br />
computationally expensive.<br />
The proposed Gaussian model with the parameter<br />
estimation scheme presented here facilitates the evaluation<br />
<strong>of</strong> <strong>hemodynamic</strong> <strong>response</strong>s at every voxel site.<br />
As demonstrated in our experiment, the concept <strong>of</strong> a<br />
global HDMF over the entire brain is incorrect. The<br />
<strong>hemodynamic</strong> <strong>response</strong> to neuronal activations is a<br />
complex function depending on the neuronal and<br />
vascular microarchitecture and the cognitive load in<br />
the region, and modulated by physiological factors<br />
such as pH, blood pressure, or metabolism. Because<br />
most <strong>of</strong> these factors may not be considered constant<br />
over the entire brain, we favored a space-variant<br />
approach. Our f<strong>MRI</strong> experiments resulted in different<br />
values <strong>of</strong> lag and dispersion <strong>for</strong> the activated regions<br />
<strong>for</strong> visual-stimulation and word-discrimination tasks,<br />
supporting our conjecture <strong>of</strong> a spatially-variant and<br />
stimulation-dependent <strong>hemodynamic</strong> <strong>response</strong> function<br />
<strong>for</strong> the brain. Although our experiments were not<br />
sufficient to make definitive conclusions about the<br />
nature <strong>of</strong> the variation <strong>of</strong> <strong>hemodynamic</strong> parameters, it<br />
became evident that they depended on a multitude <strong>of</strong><br />
factors such as brain site, stimulus condition and<br />
frequency, or individual subject. Also, the comparison<br />
<strong>of</strong> <strong>functional</strong> activities based on the same experiment<br />
with different subjects is affected by the anatomical<br />
variability <strong>of</strong> the human brain, especially at the sulcal<br />
and gyral patterns, as seen in Figure 7. In other words,<br />
the <strong>functional</strong> variability is confounded by the anatomical<br />
variability <strong>of</strong> the human brain.<br />
An exact <strong>for</strong>mulation <strong>for</strong> the degrees <strong>of</strong> freedom was<br />
presented [Worsley and Friston, 1995] to compensate<br />
<strong>for</strong> the temporal correlations present in f<strong>MRI</strong> <strong>time</strong>series.<br />
The idea <strong>of</strong> the smoothness matrix is not<br />
applicable when <strong>hemodynamic</strong>s depend both on spatial<br />
location and on stimulation conditions. There<strong>for</strong>e,<br />
the <strong>hemodynamic</strong> modulation needs to be applied to<br />
each stimulus separately. The threshold <strong>for</strong> the statistical<br />
maps to determine significant activation regions<br />
was determined empirically, considering all the data<br />
sets. There is no proper criterion <strong>for</strong> the selection <strong>of</strong><br />
this threshold, although major developments were<br />
reported [Friston et al., 1994a] in this area with the<br />
theory <strong>of</strong> random Gaussian fields. Although smoothing<br />
in both <strong>time</strong> and space improves signal-to-noise<br />
ratios <strong>of</strong> the images, it might introduce artificial temporal<br />
or spatial correlations to the images and result in<br />
poor resolution <strong>of</strong> activation maps. In our approach,<br />
the spatial and temporal effects are handled separately,<br />
but a synergetic spatiotemporal approach may be<br />
more elegant and fruitful.<br />
Univariate multiple regression <strong>analysis</strong> has been<br />
used earlier <strong>for</strong> statistical inference on significant<br />
activations with global <strong>hemodynamic</strong> parameters [Friston<br />
et al., 1995a], and an extension was presented to<br />
297