Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
Functional genomics and systems biology for studying<br />
oncogenesis<br />
Radovan Komel 1 , Petra Hudler 1 , Uro{ Raj~evi} 1 , Robert Juvan 2 ,<br />
Stanislav Rep{e 2 , Sa{a Markovi~-Predan 3<br />
1<br />
Medical Centre for Molecular Biology, Faculty of Medicine, University of Ljubljana,<br />
Ljubljana, Slovenia; 2 Clinical Department for Abdominal Surgery, Clinical Centre, Ljubljana,<br />
Slovenia; 3 Clinical Department for Gastroenterology, Clinical Centre, Ljubljana, Slovenia<br />
In contrast to traditional molecular studies of cancer that have focused on relatively<br />
small number of genes or other biomarkers where genes have generally been analyzed<br />
one or few at a time, functional genomics and systems biology are considering a cell<br />
as a system of simultaneous interactions of thousands of molecular events in a given<br />
physiological condition. The development of DNA microarray technology has made<br />
it possible to carry out large scale parallel analyses of gene expression, allowing<br />
the simultaneous comparison of the levels of expression of several thousand genes<br />
in different cell types and/or physiological conditions. The enormous progress<br />
in proteomics, enabled by recent advances in mass spectrometry combined with<br />
sequence data correlation, has enhanced speed and accuracy in identification of<br />
proteins in complex mixtures. Both approaches represent key strategies for getting<br />
complex integrative information referred as cell ‘transcriptome’ and ‘proteome’ which<br />
by bioinformatical clustering can yield systems immages relevant for diagnostics and<br />
prediction of the disease.<br />
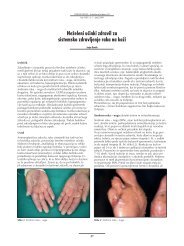
By clustering a set of 160 genes/proteins that were differentially expressed in gastric<br />
cancer as compared to normal sourounding gastric mucosa we were able to distinguish<br />
clearly pathologically trasformed cells from those not yet altered. Furthermore, by<br />
principal component analysis, cells showing the same histopathological features<br />
in two patients could be resolved in two different molecular subtypes. However,<br />
high costs of differential gene expression and proteome analysis are still preventing<br />
such studies to be carried out in a larger number of samples in order to get more<br />
generalized conclusions relevant for molecular diagnostics. It is true that attention<br />
could be paid to several genes/proteins showing prominent differences in expression,<br />
but the question remains whether gene-by-gene alteration really represent the true<br />
sequence of events in oncogenesis as proposed by the conventional oncogene<br />
activation and anti-oncogene inactivation theory. Actual poor correlation of the<br />
data obtained from transcriptome and proteome analysis as well as between<br />
different experimental platforms will be overcome in future by increased number of<br />
laboratories having access to the new technologies and thus increasing the number<br />
of cases investigated. In serach of key ‘master genes/proteins’ more attention should<br />
also be paid to overlapping a number of different physiological ‘systems’, not only<br />
‘malignant’ to the ‘normal’ cell.<br />
l43<br />
61