PDF Version - RCSB Protein Data Bank
PDF Version - RCSB Protein Data Bank
PDF Version - RCSB Protein Data Bank
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
www.pdb.org<br />
info@rcsb.org<br />
MOLECULE OF THE MONTH:<br />
CONCANAVALIN A AND<br />
CIRCULAR PERMUTATION<br />
doi: 10.2210/rcsb_pdb/mom_2010_4<br />
Evolution is a great tinkerer:<br />
once a successful plan is found,<br />
it is used again and again,<br />
often with many changes and<br />
improvements. This is easily<br />
seen in the living things around<br />
us. Most mammals have four<br />
limbs, which have evolved into<br />
all manner of arms and legs,<br />
and even into flippers and<br />
wings. Most plants are covered<br />
with leaves, which range from<br />
spiny needles to huge jungle<br />
fronds. Looking at protein<br />
sequences and structures, we see<br />
the same diversity generated<br />
through variation. <strong>Protein</strong>s<br />
evolve by slow mutation of<br />
individual amino acids, but<br />
they also evolve through larger<br />
piece-wise changes, where the<br />
genes for successful proteins are<br />
clipped and reassembled in<br />
new combinations.<br />
About the<br />
<strong>RCSB</strong> PDB Molecule of the Month<br />
Using selected molecules from the PDB<br />
archive, each feature includes an<br />
introduction to the structure and function of<br />
the molecule, a discussion of its relevance to<br />
human health and welfare, and<br />
suggestions for viewing and<br />
accessing further details.<br />
The <strong>RCSB</strong> PDB Molecule of the Month is<br />
read by students, teachers, and scientists<br />
worldwide at www.pdb.org.<br />
This April 2010 edition was written and<br />
illustrated by David S. Goodsell<br />
(<strong>RCSB</strong> PDB and The Scripps<br />
Research Institute).<br />
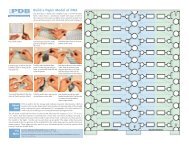
Circular Reasoning<br />
Circular permutation is one of the most unusual<br />
changes found in protein evolution. To understand<br />
where the name "circular permutation"<br />
comes from, imagine a folded protein that has the<br />
two ends of the chain near to one another. Now,<br />
chemically link these two ends. This would form<br />
a circular protein chain, with no ends. Then, clip<br />
the circularized chain in a different place. The<br />
folded protein would look the same, except that<br />
the two ends would be in a different place.<br />
Permuted <strong>Protein</strong>s<br />
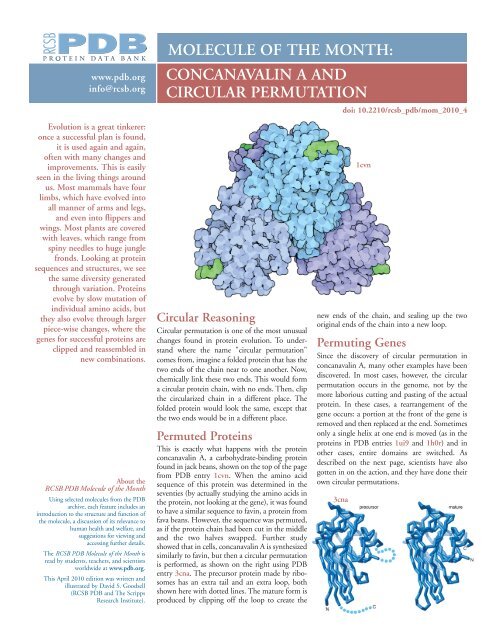
This is exactly what happens with the protein<br />
concanavalin A, a carbohydrate-binding protein<br />
found in jack beans, shown on the top of the page<br />
from PDB entry 1cvn. When the amino acid<br />
sequence of this protein was determined in the<br />
seventies (by actually studying the amino acids in<br />
the protein, not looking at the gene), it was found<br />
to have a similar sequence to favin, a protein from<br />
fava beans. However, the sequence was permuted,<br />
as if the protein chain had been cut in the middle<br />
and the two halves swapped. Further study<br />
showed that in cells, concanavalin A is synthesized<br />
similarly to favin, but then a circular permutation<br />
is performed, as shown on the right using PDB<br />
entry 3cna. The precursor protein made by ribosomes<br />
has an extra tail and an extra loop, both<br />
shown here with dotted lines. The mature form is<br />
produced by clipping off the loop to create the<br />
1cvn<br />
new ends of the chain, and sealing up the two<br />
original ends of the chain into a new loop.<br />
Permuting Genes<br />
Since the discovery of circular permutation in<br />
concanavalin A, many other examples have been<br />
discovered. In most cases, however, the circular<br />
permutation occurs in the genome, not by the<br />
more laborious cutting and pasting of the actual<br />
protein. In these cases, a rearrangement of the<br />
gene occurs: a portion at the front of the gene is<br />
removed and then replaced at the end. Sometimes<br />
only a single helix at one end is moved (as in the<br />
proteins in PDB entries 1ui9 and 1h0r) and in<br />
other cases, entire domains are switched. As<br />
described on the next page, scientists have also<br />
gotten in on the action, and they have done their<br />
own circular permutations.<br />
3cna
CONCANAVALIN A AND<br />
CIRCULAR PERMUTATION<br />
<strong>RCSB</strong> <strong>Protein</strong> <strong>Data</strong> <strong>Bank</strong><br />
The <strong>Protein</strong> <strong>Data</strong> <strong>Bank</strong> (PDB) is the<br />
single worldwide repository for the<br />
processing and distribution of 3D<br />
structure data of large molecules of<br />
proteins and nucleic acids. The <strong>RCSB</strong><br />
PDB is operated by Rutgers, The State<br />
University of New Jersey and the San<br />
Diego Supercomputer Center and the<br />
Skaggs School of Pharmacy and<br />
Pharmaceutical Sciences at the University<br />
of California, San Diego–two members<br />
of the Research Collaboratory for<br />
Structural Bioinformatics (<strong>RCSB</strong>).<br />
It is supported by funds from the<br />
National Science Foundation, the<br />
National Institute of General Medical<br />
Sciences, the Office of Science,<br />
Department of Energy, the National<br />
Library of Medicine, the National<br />
Cancer Institute, the National Institute<br />
of Neurological Disorders and Stroke<br />
and the National Institute of Diabetes<br />
& Digestive & Kidney Diseases.<br />
References:<br />
The <strong>RCSB</strong> PDB is a member of<br />
the worldwide PDB<br />
(wwPDB; www.wwpdb.org).<br />
1cvn: J.H. Naismith, R.A. Field (1996) Structural<br />
basis of trimannoside recognition by concanavalin A.<br />
J.Biol.Chem. 271: 972-976<br />
3cna: K.D. Hardman, C.F. Ainsworth (1972)<br />
Structure of concanavalin A at 2.4-A resolution.<br />
Biochemistry 11: 4910-4919<br />
1ui9: E. Inagaki, S. Kuramitsu, S. Yokoyama, M.<br />
Miyano, T.H. Tahirov; The crystal structure of chorismate<br />
mutase from thermus thermophilus. To be<br />
Published<br />
1h0r: D.A. Robinson, A.W. Roszak, M. Frederickson,<br />
C. Abell, J.R. Coggins, A.J.Lapthorn; Structural Basis<br />
for Selectivity of Oxime Based Inhibitors Towards<br />
Type II Dehydroquinase from Mycobacterium<br />
Tuberculosis. To be Published<br />
2ayh: M. Hahn, T. Keitel, U. Heinemann (1995)<br />
Crystal and molecular structure at 0.16-nm resolution<br />
of the hybrid Bacillus endo-1,3-1,4-beta-D-glucan<br />
4-glucanohydrolase H(A16-M). Eur.J.Biochem.<br />
232: 849-858<br />
1ajk, 1ajo: J. Ay, M. Hahn., K. Decanniere, K.<br />
Piotukh, R. Borriss, U. Heinemann (1998) Crystal<br />
structures and properties of de novo circularly permuted<br />
1,3-1,4-beta-glucanases. <strong>Protein</strong>s 30: 155-167<br />
1cpm: M. Hahn, K. Piotukh, R. Borriss, U.<br />
Heinemann (1994) Native-like in vivo folding of a<br />
circularly permuted jellyroll protein shown by crystal<br />
structure analysis. Proc.Natl.Acad.Sci.USA 91:<br />
10417-10421<br />
2pel: R. Banerjee, K. Das, R. Ravishankar, K. Suguna,<br />
A. Surolia, M. Vijayan (1996) Conformation, protein-carbohydrate<br />
interactions and a novel subunit<br />
association in the refined structure of peanut lectinlactose<br />
complex. J.Mol.Biol. 259: 281-296<br />
2ayh 1ajk 1ajo 1cpm<br />
Permutation on Purpose<br />
Scientists are also great tinkerers. As soon as they<br />
discovered the process of circular permutation, they<br />
had to try it out themselves. The glucanase protein<br />
shown above (PDB entries 2ayh, 1ajk, 1ajo and<br />
1cpm) has been permuted in several different ways,<br />
by cutting the gene in different places and reassembling<br />
the two pieces. In this picture, each protein<br />
chain is colored blue at one end, red at the other,<br />
and rainbow colors in between. Notice that the<br />
overall protein fold is the same in each one, except<br />
that the ends are in a different place. This demonstrates<br />
that protein folding is a robust process, and<br />
that the protein fold is determined by the sequence,<br />
even if it's shuffled around a bit.<br />
Exploring the Structure<br />
3cna<br />
2pel<br />
Circular permutation was originally discovered by<br />
comparing the amino acid sequences of concanavalin<br />
A (PDB entry 3cna) with the sequences<br />
of other lectins, such as the peanut lectin shown<br />
here (PDB entry 2pel). When the structures were<br />
solved years later, as expected, they showed very<br />
similar three-dimensional structures, in spite of<br />
the permutation of the chain. The <strong>Protein</strong><br />
Comparison Tool available in the "Compare<br />
Structures" section of the <strong>RCSB</strong> WWW site was<br />
used to overlap the proteins in this Molecule of<br />
the Month. When you use this tool, you'll find<br />
that it gets a bit confused by circularly-permuted<br />
proteins, and ends up using only part of the<br />
chain for the overlap. Fortunately, this works well<br />
enough for comparing proteins like concanavalin<br />
A and peanut lectin that have similar folds.<br />
Topics for Further Exploration<br />
1. Can you find other examples of circularlypermuted<br />
proteins in the PDB? Several<br />
databases are available to help with this,<br />
including CPDB and SISYPHUS.<br />
2. Can you think of any ways that circular<br />
permutation would improve the function<br />
of a protein?<br />
Additional reading about<br />
circular permutation<br />
U. Heinemann and M. Hahn (1995)<br />
Circular permutation of polypeptide chains:<br />
implications for protein folding and stability.<br />
Progress in Biophysics and Molecular Biology<br />
64, 121-143.<br />
D. J. Bowles and D. J. Pappin (1988) Traffic<br />
and assembly of concanavalin A. Trends in<br />
Biochemical Sciences 13, 60-64.<br />
B. A. Cunningham, J. J. Hemperly, T. P.<br />
Hopp and G. M. Edelman (1979) Favin versus<br />
concanavalin A: circularly permuted amino<br />
acid sequences. Proceedings of the National<br />
Academy of Sciences USA 76, 3218-3222.