LACTATE DEHYDROGENASE - RCSB Protein Data Bank
LACTATE DEHYDROGENASE - RCSB Protein Data Bank
LACTATE DEHYDROGENASE - RCSB Protein Data Bank
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
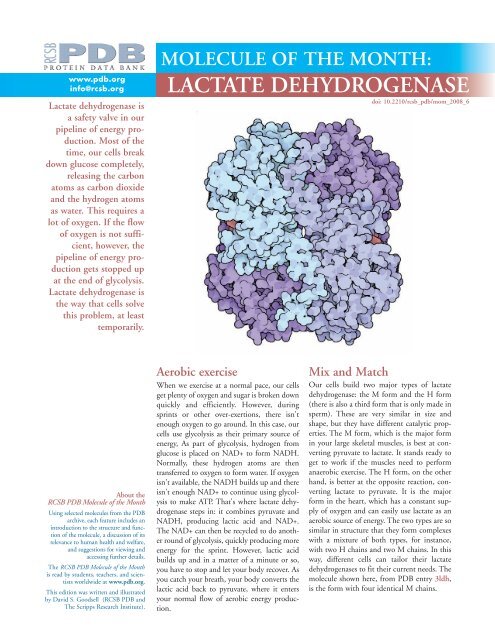
www.pdb.orginfo@rcsb.orgLactate dehydrogenase isa safety valve in ourpipeline of energy production.Most of thetime, our cells breakdown glucose completely,releasing the carbonatoms as carbon dioxideand the hydrogen atomsas water. This requires alot of oxygen. If the flowof oxygen is not sufficient,however, thepipeline of energy productiongets stopped upat the end of glycolysis.Lactate dehydrogenase isthe way that cells solvethis problem, at leasttemporarily.MOLECULE OF THE MONTH:<strong>LACTATE</strong> <strong>DEHYDROGENASE</strong>doi: 10.2210/rcsb_pdb/mom_2008_6About the<strong>RCSB</strong> PDB Molecule of the MonthUsing selected molecules from the PDBarchive, each feature includes anintroduction to the structure and functionof the molecule, a discussion of itsrelevance to human health and welfare,and suggestions for viewing andaccessing further details.The <strong>RCSB</strong> PDB Molecule of the Monthis read by students, teachers, and scientistsworldwide at www.pdb.org.This edition was written and illustratedby David S. Goodsell (<strong>RCSB</strong> PDB andThe Scripps Research Institute).Aerobic exerciseWhen we exercise at a normal pace, our cellsget plenty of oxygen and sugar is broken downquickly and efficiently. However, duringsprints or other over-exertions, there isn'tenough oxygen to go around. In this case, ourcells use glycolysis as their primary source ofenergy, As part of glycolysis, hydrogen fromglucose is placed on NAD+ to form NADH.Normally, these hydrogen atoms are thentransferred to oxygen to form water. If oxygenisn't available, the NADH builds up and thereisn't enough NAD+ to continue using glycolysisto make ATP. That's where lactate dehydrogenasesteps in: it combines pyruvate andNADH, producing lactic acid and NAD+.The NAD+ can then be recycled to do anotherround of glycolysis, quickly producing moreenergy for the sprint. However, lactic acidbuilds up and in a matter of a minute or so,you have to stop and let your body recover. Asyou catch your breath, your body converts thelactic acid back to pyruvate, where it entersyour normal flow of aerobic energy production.Mix and MatchOur cells build two major types of lactatedehydrogenase: the M form and the H form(there is also a third form that is only made insperm). These are very similar in size andshape, but they have different catalytic properties.The M form, which is the major formin your large skeletal muscles, is best at convertingpyruvate to lactate. It stands ready toget to work if the muscles need to performanaerobic exercise. The H form, on the otherhand, is better at the opposite reaction, convertinglactate to pyruvate. It is the majorform in the heart, which has a constant supplyof oxygen and can easily use lactate as anaerobic source of energy. The two types are sosimilar in structure that they form complexeswith a mixture of both types, for instance,with two H chains and two M chains. In thisway, different cells can tailor their lactatedehydrogenases to fit their current needs. Themolecule shown here, from PDB entry 3ldh,is the form with four identical M chains.
<strong>LACTATE</strong> <strong>DEHYDROGENASE</strong><strong>RCSB</strong> <strong>Protein</strong> <strong>Data</strong> <strong>Bank</strong>The <strong>Protein</strong> <strong>Data</strong> <strong>Bank</strong> (PDB) is thesingle worldwide repository for theprocessing and distribution of 3Dstructure data of large molecules ofproteins and nucleic acids. The <strong>RCSB</strong>PDB is operated by Rutgers, The StateUniversity of New Jersey and the SanDiego Supercomputer Center and theSkaggs School of Pharmacy andPharmaceutical Sciences at the Universityof California, San Diego–two membersof the Research Collaboratory forStructural Bioinformatics (<strong>RCSB</strong>).It is supported by funds from theNational Science Foundation, theNational Institute of General MedicalSciences, the Office of Science,Department of Energy, the NationalLibrary of Medicine, the NationalCancer Institute, the National Centerfor Research Resources, the NationalInstitute of Biomedical Imaging andBioengineering, the National Instituteof Neurological Disorders and Strokeand the National Institute of Diabetes& Digestive & Kidney Diseases.The <strong>RCSB</strong> PDB is a member ofthe worldwide PDB(wwPDB; www.wwpdb.org).Additional ReadingD.L. Nelson, M.M. Cox (2000) LehningerPrinciples of Biochemistry. Worth Publishers.M. T. Madigan, J. M. Martinko, J. Parker(2000) Brock Biology of Microorganisms.Prentice Hall.J.A. Read, K.W. Wilkinson, R. Tranter, R.B.Sessions, R.L. Brady (1999) Chloroquinebinds in the cofactor binding site ofPlasmodium falciparum lactate dehydrogenase.Journal of Biological Chemistry 274: 10213-10218.S. Iwata, K. Kamata, S. Yoshida, T. Minowa,T. Ohta (1994) T and R states in the crystalsof bacterial L-lactate dehydrogenase reveal themechanism for allosteric control. Structure 1:176-185.J.J. Holbrook, A. Liljas, S.J. Steindel andM.G. Rossman (1975) LactateDehydrogenase. In The Enzymes P. D. Boyer,editor. Academic Press. Volume XI, pages191-292.FermentationSome bacteria obtain most of their energy by conversion of glucose into lactose. This process iscalled fermentation, and you have probably experienced the results of it at the dinner table.Fermenting bacteria are used to convert milk into yogurt and fermented lactic acid is an importantpart of the sharp flavors of sauerkraut and sourdough bread. The bacterial lactose dehydrogenaseshown here is an allosteric enzyme. Binding of fructose 1,6-bisphosphate, one of themolecules formed in the early steps of glycolysis, causes the enzyme to change into an activeshape. Remarkably, PDB entry 1lth, shown here, contains two separate enzyme molecules, onein the active state (left) and one in the inactive state (right).Exploring the StructureThe protozoan parasites that cause malaria arethought to rely on glycolysis for most of their energyduring part of their cycle of infection.Researchers are now looking for drugs to block theaction of lactate dehydrogenase as a way of attackingthese parasites and curing the infection. Thestructure shown here (PDB entry 1cet) has fourmolecules of chloroquine bound in the active sitesof the lactate dehydrogenase found in thePlasmodium parasite. Chloroquine is one of themajor drugs used to treat malaria, however, itsmajor site of action probably isn't at this enzyme;instead, it is thought to block the unusual methodsthat the parasite must use to feed on blood. But researchers are exploring many other antimalarialmolecules that target lactate dehydrogenase, as seen in other PDB entries such as 1t24,1t25, and similar structures.References:3ldh: White, J.L., Hackert, M.L., Buehner, M., Adams, M.J., Ford, G.C., Lentz Jr., P.J., Smiley, I.E., Steindel, S.J., Rossmann,M.G. (1976) A comparison of the structures of apo dogfish M4 lactate dehydrogenase and its ternary complexes. J.Mol.Biol. 102:759-779.1lth: S. Iwata, K. Kamata, S. Yoshida, T. Minowa, T. Ohta (1994) T and R states in the crystals of bacterial L-lactate dehydrogenasereveal the mechanism for allosteric control. Nat.Struct.Biol. 1: 176-185.1t24: A. Cameron, J. Read, R. Tranter, V.J. Winter, R.B. Sessions, R.L. Brady, L. Vivas, A. Easton, H. Kendrick, S.L. Croft, D. Barros,J.L. Lavandera, J.J. Martin, F. Risco, S. Garcia-Ochoa, F.J. Gamo, L. Sanz, L. Leon, J.R. Ruiz, R. Gabarro, A. Mallo, F.G. de las Heras(2004) Identification and activity of a series of azole-based compounds with lactate dehydrogenase-directed anti-malarial activity.J.Biol.Chem. 279: 31429-31439.