Haematologica 2000;85:supplement to no. 10 - Supplements ...
Haematologica 2000;85:supplement to no. 10 - Supplements ...
Haematologica 2000;85:supplement to no. 10 - Supplements ...
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
<strong>10</strong><br />
J. Oldenburg et al.<br />
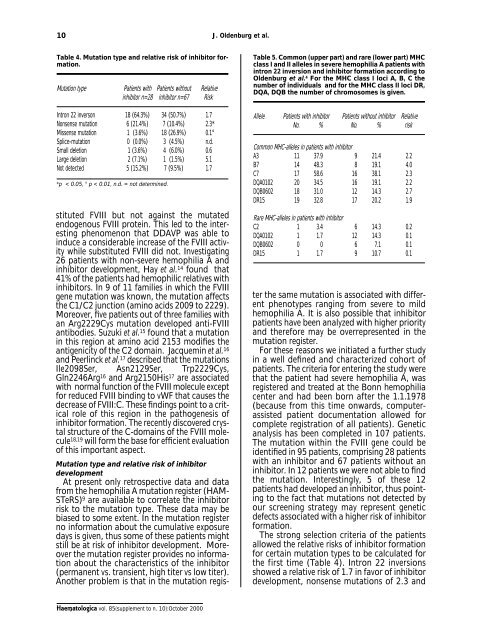
Table 4. Mutation type and relative risk of inhibi<strong>to</strong>r formation.<br />
Mutation type Patients with Patients without Relative<br />
inhibi<strong>to</strong>r n=28 inhibi<strong>to</strong>r n=67 Risk<br />
Intron 22 inverson 18 (64.3%) 34 (50.7%) 1.7<br />
Nonsense mutation 6 (21.4%) 7 (<strong>10</strong>.4%) 2.3*<br />
Missense mutation 1 (3.6%) 18 (26.9%) 0.1°<br />
Splice-mutation 0 (0.0%) 3 (4.5%) n.d.<br />
Small deletion 1 (3.6%) 4 (6.0%) 0.6<br />
Large deletion 2 (7.1%) 1 (1.5%) 5.1<br />
Not detected 5 (15.2%) 7 (9.5%) 1.7<br />
*p < 0.05, ° p < 0.01, n.d. = <strong>no</strong>t determined.<br />
stituted FVIII but <strong>no</strong>t against the mutated<br />
endoge<strong>no</strong>us FVIII protein. This led <strong>to</strong> the interesting<br />
phe<strong>no</strong>me<strong>no</strong>n that DDAVP was able <strong>to</strong><br />
induce a considerable increase of the FVIII activity<br />
while substituted FVIII did <strong>no</strong>t. Investigating<br />
26 patients with <strong>no</strong>n-severe hemophilia A and<br />
inhibi<strong>to</strong>r development, Hay et al. 14 found that<br />
41% of the patients had hemophilic relatives with<br />
inhibi<strong>to</strong>rs. In 9 of 11 families in which the FVIII<br />
gene mutation was k<strong>no</strong>wn, the mutation affects<br />
the C1/C2 junction (ami<strong>no</strong> acids 2009 <strong>to</strong> 2229).<br />
Moreover, five patients out of three families with<br />
an Arg2229Cys mutation developed anti-FVIII<br />
antibodies. Suzuki et al. 15 found that a mutation<br />
in this region at ami<strong>no</strong> acid 2153 modifies the<br />
antigenicity of the C2 domain. Jacquemin et al. 16<br />
and Peerlinck et al. 17 described that the mutations<br />
Ile2098Ser, Asn2129Ser, Trp2229Cys,<br />
Gln2246Arg 16 and Arg2150His 17 are associated<br />
with <strong>no</strong>rmal function of the FVIII molecule except<br />
for reduced FVIII binding <strong>to</strong> vWF that causes the<br />
decrease of FVIII:C. These findings point <strong>to</strong> a critical<br />
role of this region in the pathogenesis of<br />
inhibi<strong>to</strong>r formation. The recently discovered crystal<br />
structure of the C-domains of the FVIII molecule<br />
18,19 will form the base for efficient evaluation<br />
of this important aspect.<br />
Table 5. Common (upper part) and rare (lower part) MHC<br />
class I and II alleles in severe hemophilia A patients with<br />
intron 22 inversion and inhibi<strong>to</strong>r formation according <strong>to</strong><br />
Oldenburg et al. 4 For the MHC class I loci A, B, C the<br />
number of individuals and for the MHC class II loci DR,<br />
DQA, DQB the number of chromosomes is given.<br />
Allele Patients with inhibi<strong>to</strong>r Patients without inhibi<strong>to</strong>r Relative<br />
No. % No. % risk<br />
Common MHC-alleles in patients with inhibi<strong>to</strong>r<br />
A3 11 37.9 9 21.4 2.2<br />
B7 14 48.3 8 19.1 4.0<br />
C7 17 58.6 16 38.1 2.3<br />
DQA0<strong>10</strong>2 20 34.5 16 19.1 2.2<br />
DQB0602 18 31.0 12 14.3 2.7<br />
DR15 19 32.8 17 20.2 1.9<br />
Rare MHC-alleles in patients with inhibi<strong>to</strong>r<br />
C2 1 3.4 6 14.3 0.2<br />
DQA0<strong>10</strong>2 1 1.7 12 14.3 0.1<br />
DQB0602 0 0 6 7.1 0.1<br />
DR15 1 1.7 9 <strong>10</strong>.7 0.1<br />
Mutation type and relative risk of inhibi<strong>to</strong>r<br />
development<br />
At present only retrospective data and data<br />
from the hemophilia A mutation register (HAM-<br />
STeRS) 9 are available <strong>to</strong> correlate the inhibi<strong>to</strong>r<br />
risk <strong>to</strong> the mutation type. These data may be<br />
biased <strong>to</strong> some extent. In the mutation register<br />
<strong>no</strong> information about the cumulative exposure<br />
days is given, thus some of these patients might<br />
still be at risk of inhibi<strong>to</strong>r development. Moreover<br />
the mutation register provides <strong>no</strong> information<br />
about the characteristics of the inhibi<strong>to</strong>r<br />
(permanent vs. transient, high titer vs low titer).<br />
A<strong>no</strong>ther problem is that in the mutation register<br />
the same mutation is associated with different<br />
phe<strong>no</strong>types ranging from severe <strong>to</strong> mild<br />
hemophilia A. It is also possible that inhibi<strong>to</strong>r<br />
patients have been analyzed with higher priority<br />
and therefore may be overrepresented in the<br />
mutation register.<br />
For these reasons we initiated a further study<br />
in a well defined and characterized cohort of<br />
patients. The criteria for entering the study were<br />
that the patient had severe hemophilia A, was<br />
registered and treated at the Bonn hemophilia<br />
center and had been born after the 1.1.1978<br />
(because from this time onwards, computerassisted<br />
patient documentation allowed for<br />
complete registration of all patients). Genetic<br />
analysis has been completed in <strong>10</strong>7 patients.<br />
The mutation within the FVIII gene could be<br />
identified in 95 patients, comprising 28 patients<br />
with an inhibi<strong>to</strong>r and 67 patients without an<br />
inhibi<strong>to</strong>r. In 12 patients we were <strong>no</strong>t able <strong>to</strong> find<br />
the mutation. Interestingly, 5 of these 12<br />
patients had developed an inhibi<strong>to</strong>r, thus pointing<br />
<strong>to</strong> the fact that mutations <strong>no</strong>t detected by<br />
our screening strategy may represent genetic<br />
defects associated with a higher risk of inhibi<strong>to</strong>r<br />
formation.<br />
The strong selection criteria of the patients<br />
allowed the relative risks of inhibi<strong>to</strong>r formation<br />
for certain mutation types <strong>to</strong> be calculated for<br />
the first time (Table 4). Intron 22 inversions<br />
showed a relative risk of 1.7 in favor of inhibi<strong>to</strong>r<br />
development, <strong>no</strong>nsense mutations of 2.3 and<br />
<strong>Haema<strong>to</strong>logica</strong> vol. <strong>85</strong>(<strong>supplement</strong> <strong>to</strong> n. <strong>10</strong>):Oc<strong>to</strong>ber <strong>2000</strong>