The three-dimensional structure of humic substances and soil ...
The three-dimensional structure of humic substances and soil ...
The three-dimensional structure of humic substances and soil ...
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
First, a preliminary 2 D <strong>structure</strong> <strong>of</strong> the C-C skeleton<br />
is drawn by h<strong>and</strong> into the workspace using the drawing<br />
tool <strong>and</strong> is far from perfect as shown in Fig. 3. In addi-<br />
tion to carbon, the elements oxygen <strong>and</strong> nitrogen are in-<br />
cluded (Default Element). In this manner 308 carbon-,<br />
90 oxygen- <strong>and</strong> 5 nitrogen-atoms (403 atoms in total) are<br />
positioned.<br />
Second, hydrogen is attached with accurate bond<br />
lengths <strong>and</strong> angles to the preliminary <strong>structure</strong> using the<br />
s<strong>of</strong>tware (Add Hydrogen) as shown in Fig. 4. Whereas in<br />
the original publication the 2 D HA subunit <strong>structure</strong> [2]<br />
was incomplete <strong>and</strong> was supposed to have a wide variety<br />
<strong>of</strong> different bonding sequences (see :k-- symbols), the mol-<br />
ecule in Fig. 4 represents a defined, intact HA monomer<br />
(738 atoms). Thus, in order to obtain a complete chemi-<br />
cal compound, it had to be supplemented by 7 hydrogen<br />
atoms. <strong>The</strong> corresponding elemental composition then is<br />
C308H335090N 5 with a molecular mass <strong>of</strong> 5547.004g<br />
mol -~ <strong>and</strong> an elemental analysis <strong>of</strong> 66.69% C, 6.09% H,<br />
25.96% O, <strong>and</strong> 1.26% N.<br />
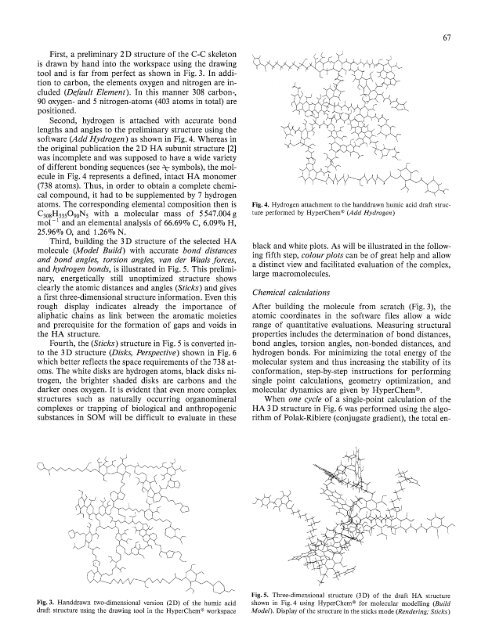
Third, building the 3 D <strong>structure</strong> <strong>of</strong> the selected HA<br />
molecule (Model BuiM) with accurate bond distances<br />
<strong>and</strong> bond angles, torsion angles, van der Waals forces,<br />
<strong>and</strong> hydrogen bonds, is illustrated in Fig. 5. This prelimi-<br />
nary, energetically still unoptimized <strong>structure</strong> shows<br />
clearly the atomic distances <strong>and</strong> angles (Sticks) <strong>and</strong> gives<br />
a first <strong>three</strong>-<strong>dimensional</strong> <strong>structure</strong> information. Even this<br />
rough display indicates already the importance <strong>of</strong><br />
aliphatic chains as link between the aromatic moieties<br />
<strong>and</strong> prerequisite for the formation <strong>of</strong> gaps <strong>and</strong> voids in<br />
the HA <strong>structure</strong>.<br />
Fourth, the (Sticks) <strong>structure</strong> in Fig. 5 is converted in-<br />
to the 3 D <strong>structure</strong> (Disks, Perspective) shown in Fig. 6<br />
which better reflects the space requirements <strong>of</strong> the 738 at-<br />
oms. <strong>The</strong> white disks are hydrogen atoms, black disks ni-<br />
trogen, the brighter shaded disks are carbons <strong>and</strong> the<br />
darker ones oxygen. It is evident that even more complex<br />
<strong>structure</strong>s such as naturally occurring organomineral<br />
complexes or trapping <strong>of</strong> biological <strong>and</strong> anthropogenic<br />
<strong>substances</strong> in SOM will be difficult to evaluate in these<br />
Fig. 3. H<strong>and</strong>drawn two-<strong>dimensional</strong> version (2D) <strong>of</strong> the <strong>humic</strong> acid<br />
draft <strong>structure</strong> using the drawing tool in the HyperChem ® workspace<br />
-2<br />
Fig. 4. Hydrogen attachment to the h<strong>and</strong>drawn <strong>humic</strong> acid draft struc-<br />
ture performed by HyperChem ® (Add Hydrogen)<br />
black <strong>and</strong> white plots. As will be illustrated in the follow-<br />
ing fifth step, colourplots can be <strong>of</strong> great help <strong>and</strong> allow<br />
a distinct view <strong>and</strong> facilitated evaluation <strong>of</strong> the complex,<br />
large macromolecules.<br />
Chemical calculations<br />
After building the molecule from scratch (Fig. 3), the<br />
atomic coordinates in the s<strong>of</strong>tware files allow a wide<br />
range <strong>of</strong> quantitative evaluations. Measuring structural<br />
properties includes the determination <strong>of</strong> bond distances,<br />
bond angles, torsion angles, non-bonded distances, <strong>and</strong><br />
hydrogen bonds. For minimizing the total energy <strong>of</strong> the<br />
molecular system <strong>and</strong> thus increasing the stability <strong>of</strong> its<br />
conformation, step-by-step instructions for performing<br />
single point calculations, geometry optimization, <strong>and</strong><br />
molecular dynamics are given by HyperChem ®.<br />
When one cycle <strong>of</strong> a single-point calculation <strong>of</strong> the<br />
HA 3 D <strong>structure</strong> in Fig. 6 was performed using the algo-<br />
rithm <strong>of</strong> Polak-Ribiere (conjugate gradient), the total en-<br />
Fig. 5. Three-<strong>dimensional</strong> <strong>structure</strong> (3D) <strong>of</strong> the draft HA <strong>structure</strong><br />
shown in Fig. 4 using HyperChem ® for molecular modelling (Build<br />
Model). Display <strong>of</strong> the <strong>structure</strong> in the sticks mode (Rendering; Sticks)<br />
67





![Legislativa - Označování potravin [režim kompatibility]](https://img.yumpu.com/15533670/1/190x135/legislativa-oznacovani-potravin-rezim-kompatibility.jpg?quality=85)







