Protocols for field and laboratory rodent studies - HAL
Protocols for field and laboratory rodent studies - HAL
Protocols for field and laboratory rodent studies - HAL
- No tags were found...
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
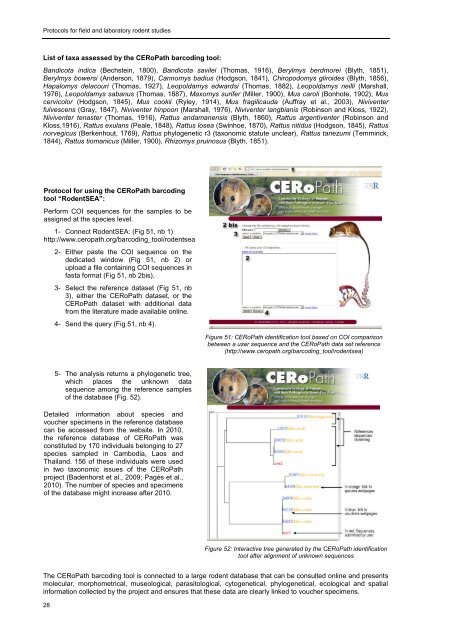
<strong>Protocols</strong> <strong>for</strong> <strong>field</strong> <strong>and</strong> <strong>laboratory</strong> <strong>rodent</strong> <strong>studies</strong>List of taxa assessed by the CERoPath barcoding tool:B<strong>and</strong>icota indica (Bechstein, 1800), B<strong>and</strong>icota savilei (Thomas, 1916), Berylmys berdmorei (Blyth, 1851),Berylmys bowersi (Anderson, 1879), Cannomys badius (Hodgson, 1841), Chiropodomys gliroides (Blyth, 1856),Hapalomys delacouri (Thomas, 1927), Leopoldamys edwardsi (Thomas, 1882), Leopoldamys neilli (Marshall,1976), Leopoldamys sabanus (Thomas, 1887), Maxomys surifer (Miller, 1900), Mus caroli (Bonhote, 1902), Muscervicolor (Hodgson, 1845), Mus cookii (Ryley, 1914), Mus fragilicauda (Auffray et al., 2003), Niviventerfulvescens (Gray, 1847), Niviventer hinpoon (Marshall, 1976), Niviventer langbianis (Robinson <strong>and</strong> Kloss, 1922),Niviventer tenaster (Thomas, 1916), Rattus <strong>and</strong>amanensis (Blyth, 1860), Rattus argentiventer (Robinson <strong>and</strong>Kloss,1916), Rattus exulans (Peale, 1848), Rattus losea (Swinhoe, 1870), Rattus nitidus (Hodgson, 1845), Rattusnorvegicus (Berkenhout, 1769), Rattus phylogenetic r3 (taxonomic statute unclear), Rattus tanezumi (Temminck,1844), Rattus tiomanicus (Miller, 1900), Rhizomys pruinosus (Blyth, 1851).Protocol <strong>for</strong> using the CERoPath barcodingtool “RodentSEA”:Per<strong>for</strong>m COI sequences <strong>for</strong> the samples to beassigned at the species level.1- Connect RodentSEA: (Fig 51, nb 1)http://www.ceropath.org/barcoding_tool/<strong>rodent</strong>sea2- Either paste the COI sequence on thededicated window (Fig 51, nb 2) orupload a file containing COI sequences infasta <strong>for</strong>mat (Fig 51, nb 2bis).3- Select the reference dataset (Fig 51, nb3), either the CERoPath dataset, or theCERoPath dataset with additional datafrom the literature made available online.4- Send the query (Fig 51, nb 4).Figure 51: CERoPath identification tool based on COI comparisonbetween a user sequence <strong>and</strong> the CERoPath data set reference(http://www.ceropath.org/barcoding_tool/<strong>rodent</strong>sea)5- The analysis returns a phylogenetic tree,which places the unknown datasequence among the reference samplesof the database (Fig. 52).Detailed in<strong>for</strong>mation about species <strong>and</strong>voucher specimens in the reference databasecan be accessed from the website. In 2010,the reference database of CERoPath wasconstituted by 170 individuals belonging to 27species sampled in Cambodia, Laos <strong>and</strong>Thail<strong>and</strong>. 156 of these individuals were usedin two taxonomic issues of the CERoPathproject (Badenhorst et al., 2009; Pagès et al.,2010). The number of species <strong>and</strong> specimensof the database might increase after 2010.Figure 52: Interactive tree generated by the CERoPath identificationtool after alignment of unknown sequencesThe CERoPath barcoding tool is connected to a large <strong>rodent</strong> database that can be consulted online <strong>and</strong> presentsmolecular, morphometrical, museological, parasitological, cytogenetical, phylogenetical, ecological <strong>and</strong> spatialin<strong>for</strong>mation collected by the project <strong>and</strong> ensures that these data are clearly linked to voucher specimens.28