Tutorial for the WGCNA package for R - UCLA Human Genetics
Tutorial for the WGCNA package for R - UCLA Human Genetics
Tutorial for the WGCNA package for R - UCLA Human Genetics
- No tags were found...
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
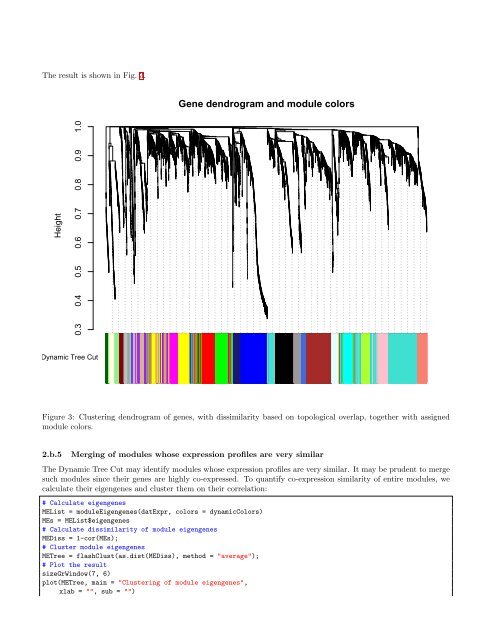
The result is shown in Fig. 3.Gene dendrogram and module colorsHeight0.3 0.4 0.5 0.6 0.7 0.8 0.9 1.0Dynamic Tree Cutas.dist(dissTom)hclust (*, "average")Figure 3: Clustering dendrogram of genes, with dissimilarity based on topological overlap, toge<strong>the</strong>r with assignedmodule colors.2.b.5Merging of modules whose expression profiles are very similarThe Dynamic Tree Cut may identify modules whose expression profiles are very similar. It may be prudent to mergesuch modules since <strong>the</strong>ir genes are highly co-expressed. To quantify co-expression similarity of entire modules, wecalculate <strong>the</strong>ir eigengenes and cluster <strong>the</strong>m on <strong>the</strong>ir correlation:# Calculate eigengenesMEList = moduleEigengenes(datExpr, colors = dynamicColors)MEs = MEList$eigengenes# Calculate dissimilarity of module eigengenesMEDiss = 1-cor(MEs);# Cluster module eigengenesMETree = flashClust(as.dist(MEDiss), method = "average");# Plot <strong>the</strong> resultsizeGrWindow(7, 6)plot(METree, main = "Clustering of module eigengenes",xlab = "", sub = "")