Chapter 6 The polyatomic molecules
Chapter 6 The polyatomic molecules
Chapter 6 The polyatomic molecules
- No tags were found...
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
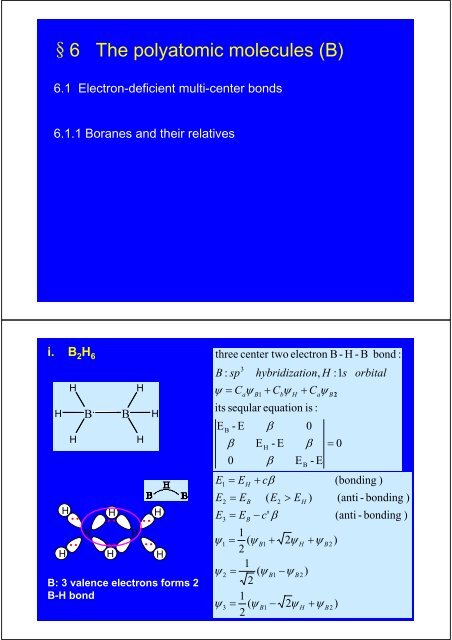
§6 <strong>The</strong> <strong>polyatomic</strong> <strong>molecules</strong> (B)6.1 Electron-deficient multi-center bonds6.1.1 Boranes and their relativesi. B 2 H 6HHHHHBBHHBBB: 3 valence electrons forms 2B-H bondHHHHHthree center two electron B - H - BB : spits seqular equation is :EB03- Ehybridization,H :1sψ = CaψB1+ CbψH+ CaψB23123βE = EEE12ψ =ψψ= E= E==HBBEβHβ+ cβ1( ψB1+21( ψB121( ψB1−2- E( E2− c'βE> E2ψ−ψB22ψHHBH0β))- E+ ψ+ ψ= 0B2B2))orbital(bonding )bond :(anti - bonding )(anti - bonding )
Three center two electron B-H-B bond:ψ 3ψ B1 ψ B 2ψ 2ψ Hψ 1HHBHHBHHii. B 5 H 9Electron-deficient multi-center bondsHBHBHHHBHHBHBHEBβ- E β βEBβ β- E β = 0EB- Esolve seqular equationEE12= E= EB3+ 2β= E1ψ1= ( ψa3B− β+ ψ + ψ )baBBB3center-2electron bonds
Chemical bonds in Boranes(a) Single bondB-HB-B(b) 3center-2electron bonds(c) Other polycenter-polyelectron bondElectron-deficient multi-center bonds6.1.2 For B nH n+m’s open structure (including to nido and arachno)n equals to number of B-H bondsIts stationary condition:x = m-st = n-sy = (2s-m)/2p sets of styxp isomers<strong>The</strong> topological structure of B 6 H 10B n H n+m (WilliamLipscomb) Nobelprize,1976BHHBHHBHBBHHBBHHHBBHBHBs t y x(3 isomers)HHBBHBBBHHBHBBHHHHHBBHHBBH 2HBBH 2BHH(4220) (3311) (2402)
Structure of BoranesB 4 H 10B 5 H 9 B 6 H 10B 8 H 12 B 10 H 14Electron-deficient multi-center bonds6.1.3 For Borohydride ions B nH n2-’s closo structure andcarboranes C n B n H n+n’+mB n H n2-(closo)B 4 H2-4 ( tetrahedral)B 6 H2-6 (octahedral)B 8 H2-8 (dodecahedral)B 12 H2-12 (icosahedral)Electron-deficient multi-center bonds
polyhedra with n vertices“Closo” series -formula B n H n2-Total valence electrons (CVE) = 3n (from B) +n (from H)+2 (negative charge) = 4n+2• Each BH unit uses 2 electrons. Hence skeletal orframework electrons (NFE=4n+2-2n=2n+2)• <strong>The</strong>re is little tendency to add H+ and form neutral species.<strong>The</strong> closo species are , in effect, the anions of quite strongacids.• Structures are those of the appropriate polyhedra with nvertices.
“Nido” series – formula B n H (n+4)•Total valence electrons(VEC) = 3n (B) +n(H) + 4 (extra H and/or negative charges)= 4n +4•Framework electrons (NFE) =2n+4 (n+2pairs).•<strong>The</strong> structure of the “nido” compound isbased on the “closo” polyhedron with onemore vertex than the “nido” compound.“Arachno” series – formula B n H (n+6)•Total valence electrons(VEC) = 4n +6•Framework electrons (NFE) =2n+4 (n+2pairs).•<strong>The</strong> structure of the “nido” compound isbased on the “closo” polyhedron with twomore vertex than the “nido” compound.Electron-deficient multi-center bonds6.1.4 other deficient electron compoundsElectron-deficient multi-center bondsBoron groupB、Al、Ga、In、TlH 3 C120º AlH 3 CCH 3 CH 370ºAlC CH 3H 32.14 Å1.94ÅGaseity: monomerSolid state: polymer
Alkali metals and alkali earth metals[Li(CH 3 )] 4H 3 CBLiCH 3Be Be CH 3 H 3 C Be Be Be CH 3BeCH 3HBe(CH 3) 2H 3H 3 H 3CC CCH 3CH 3HHHH 3CCH 3BeBeHHH 3CCH 3BBeHHH 3CCH 3Bedimertrimerpolymer6.2 Chemical bonds in the coordination compounds
Coordination compoundscompounds composed of a metal atom or ion and one ormore ligands.– Ligands usually donate electrons to the metal– Includes organometallic compoundsNew theories arose to describe bonding.–Valence bond, crystal field, and ligand field.OOOCH 2 CCH 2CON CH 2 CH 2 NOCH 2 CCH 2COOOethylenediaminetetraacetate (EDTA 4– )(hexadentate)
6.2.1 Coordination polyhedronmolecualrAg(NH 3 ) 2+C.N.2hybridizationtypespSymmetrygeometrylinearCuCl 3-3sp 2D 3htriangularNi(CO) 44sp 3T dtetrahedralPtCl 42-4dsp 2D 4hsquare planarFe(CO) 555dsp 3d 2 sp 2D 3hC 4vTrigonalbipyramidalsquarepyramidFeF 64-6d 2 sp 3O hoctahedralothers8D 4htetragonal810D 4dAnti-squarepyramidBicapped squareantiprism12I hicosahedral6.2.2 Crystal Field Model• focuses on the energies of the d orbitals.• Assumptions• 1. Ligands are negative point charges.• 2. Metal-ligand bonding is entirely ionic.• strong-field (low-spin): large splitting of d orbitals• weak-field (high-spin): small splitting of d orbitals
Crystal-Field <strong>The</strong>oryA. Crystal field splittingCrystal-Field <strong>The</strong>oryA. Crystal field splittingd z2d x-y 2 2e gcrystal fieldsplitting, ΔE3dd xy d xz d yzt 2g"naked"metalcoordinationcomplex
A. Crystal field splitting1. spectrochemical seriesCN – > NO 2– > en > NH 3 > H 2 O > C 2 O 42–> OH – > F – >Cl – > Br – > I –strongest bondlargest Δabsorbs appearsweakest Ni(H 2 O)2+6
II. Crystal-Field <strong>The</strong>oryB. Magnetic propertiese.g., Fe(NH 3 ) 62+(Fe 2+ = d 6 )ore gt 2ge gt 2glow-spin complex(maximum pairing)diamagnetichigh-spin complex(minimum pairing)paramagneticfoundexperimentallyII. Crystal-Field <strong>The</strong>oryB. Magnetic propertiesCompetition between: crystal field splitting (Δ)electron pairing energy (P)when Δ < P ⇒ high-spin complexwhen Δ > P ⇒ low-spin complexGenerally: d 1 , d 2 , d 3 : always high-spind 4 , d 6 : high-spin with ligands ≤ H 2 Olow-spin with ligands > H 2 Od 5 : high-spin with all ligands except CN –d 7 -d 10always low-spin
II. Crystal-Field <strong>The</strong>oryB. Magnetic propertiese.g., [Cr(H 2 O) 6 ] 3+e.g., [Ni(NH 3 ) 6 ] 2+e.g., [Fe(H 2 O) 6 ] 3+ vs [Fe(CN) 6 ] 3–6.2.3 σ ligands and σ bond345Categories of central metal valence orbitals:1σ group: s, p x ,p y ,p z , d x 2 -y 2, d z 262π group: d xy ,d xz ,d yz
6.2.3 σ ligands and σ bond1ψ1 = ϕ4s± ( σ1+ σ2 + σ3 + σ4 + σ5 + σ6)61 1 1ψ2= ϕ4px± ( σ1−σ4) ψ3= ϕ4py± ( σ2 −σ5) ψ4= ϕ4pz± ( σ3−σ6)2221ψ5= ϕ 2 ±3 (2 σ3+ 2 σdz6−σ1−σ2 −σ4 −σ5)2 3ψ 16= ϕ 2 2 ±3dxy (1 2 4 5 )2σ − σ + σ − σ−
Energetic diagram of σ molecular orbitals6.2.4 π − Bonding• A π-donor ligand donates electrons to the metalcenter in an interaction that involves a filled ligandorbital and an empty metal orbital.– Cl-, Br-, and I- donates p electrons to the metal center• A π-acceptor ligand accepts electrons from themetal center in an interaction that involves a filledmetal orbital and an empty ligand orbital.– CO, N 2 , NO, and alkenes accept electrons into theirvacant anti-bonding MO’s.π-acceptor ligands can stabilize low oxidation statemetal complexes.
Ni(CO) 4[PtCl 3 (C 2 H 4 )] -Ni C O π* bondσ bond (d xy , d xz , d yz )ClClClPtHHCCHHCO HOMOPtCπ bonding orbitalCCO LUMOπ anti-bonding orbitald - π conjugationCO bond made weakened6.2.5 18-electron rule• A low oxidation state organometallic complex contains π-acceptor ligands and the metal center tends to acquire 18electrons in its valence shell.• Rules:– Treat ligands as neutral entities.– <strong>The</strong> number of valence electrons for a zero-valentmetal center is equal to the group number.–Eg: Cr (group 6) in Cr(CO) 6 , Fe (group 8) in Fe(CO) 5 ,and Ni (group 10) in Ni(CO) 4
18-electron rule• Many ligands donate more than 1 electron.1-electron donor: H • (in any bonding mode), and terminal Cl • ,Br • , I • ,R • (e.g. R=alkyl or Ph) or RO • ;2-electron donor: CO, PR 3 , P(OR) 3 , R 2 C=CR 2 (η 2 -alkene),R 2 C: (carbene)3-electron donor: η 3 –C 3 H 5• (allyl radical), RC (carbyne), μ- Cl • ,μ- Br • , μ- I • , μ- R 2 P • ;4-electron donor: η 4 –diene, η 4 –C 4 R 4 (cyclo-butadienes);5-electron donor: η 5 –C 5 H 5• , μ 3 -Cl • , μ 3 -Br • , μ 3 -I • , μ 3 -R 2 P • ;6-electron donor: η 6 –C 6 H 6 , η 6 –C 6 H 5 Me;1- or 3-electron donor: NOExample: Ferrocene6.3 Prediction of structural features ofinorganic iono-covalent compoundswith tetrahedral anion complexes
%O T and %O [3] and %O [4] = 0Hume-Rothery’s 8 - N rule (1930/31)Rule rationalizes observed structural featuresof (post-transition) main group elements. Byforming the correct number of shared bondswith its neighbors each atom succeeds tocomplete its octet.<strong>The</strong> number of bonds of an element is8 - N where N is its column number in theperiodic table (only for 4 ≤ N ≤ 8).Element structures which obey Hume-Rothery’s 8 - N ruleN = 44 bondsN = 53 bondsN = 62 bondsC D , Si, Ge, α-Sn P, As, Sb, Bi S, Se, TeN = 71 bondN = 80 bondsF, Cl, Br, I He, Ne, Ar, Kr, Xe, Rn
Generalized 8 - N rulePearson (1964), Hulliger & Mooser (1965)8 - VEC A = AA - CC / (n/m) for C m A nVEC A : Number of valence electrons par anionVEC A < 8, AA>0, CC=0 Polyanionic val. comp.VEC A = 8, AA=0, CC=0 Normal valence compoundVEC A > 8, AA=0, CC>0 Polycationic val. comp.AA: Average number of A-A bonds per anionCC; Average number of C-C bonds per cations and/orelectrons used for inert-electron pairs on cations
Polyanionic valence compound (1)K 6 Pd 2+ Se 20 : VEC A = 128/20, AA=8/5K 6 Pd[Se 5 ] 4AA = 1AA = 4/3AA = 6/4AA = 8/5N‘ A/M = 2/(2 - AA)N‘ A/M : Averagenumber of atoms in anon-cyclic chargedanion moleculePolyanionic valence compounds (2)• LaAs 2 : VEC A = 6.5; AA = 3/2; N A/M = 4• La 2^[As 4 ] LT, La 4^[As 3 ] ^[As 5 ] HT• CsTe 4 : VEC A = 6.25; AA = 7/4; N A/M = 8• Cs 2^[Te 8 ]• Th 2 S 5 : VEC A = 7.6; AA = 2/5• Th 2^[S 2 ][S] 3• Sr 5 Si 3 : VEC A = 7.33; AA = 2/3• Sr 5^[S 2 ][S]
Polycationic valence compounds• HgCl : VEC A = 9; CC = 1• [Hg-Hg]Cl 2• CCl 3 : VEC A = 8.33; CC = 1• [C-C]Cl 6• SiAs : VEC A = 9; CC = 1• [Si-Si]As 26.4 Transition-metal cluster compounds6.4.1 Metal-metal bondL n M-ML n , L n M=ML n , L n M≡ML n ,… so called clusterRe 2 Cl 82-Re, d 5 s 2Re-Re 2.24Å (2.76 Å in Re crystal) Cl…Cl 3.32 ÅRe 3+ , d 4 , dsp 2 hybridization( σ bond), remain four d and one p orbitalσ ( d − d 2)π ( dπ ( dδ ( dz2xzyzxy− d− d− dzxzyzxy)))Quadruple Bond2 4 2σ π δAnalog: Mo 2 (O 2 CR) 4 and Cr 2 (O 2 CR) 4[Re2Cl8] 2-
6.4 Transition-metal cluster compounds6.4.1 Metal-metal bondL n M-ML n , L n M=ML n , L n M≡ML n ,… so called clusterIr(CO) 3IrIr(CO) 3Ir(CO) 3(CO) 3Ir 4 (CO) 126.4.2 Cluster geometryi. Structural polyhedron
ii.Electron countingb=12(18n−g)b: bond valence (total number of metal-metal bonds)n: number of metal atomsg: total electrons in valence shell, including allIn the case that main group atoms are included:b=12(18n1 + 8n2− g)
iii. Bond valence and the cluster geometryb=12(18n−g)Tri-nuclear compoundsMetal cluster compoundsOs 3(CO) 9(μ 3-S) 2g50b2M-M/pmOs-Os,281.3OsOsOsMn 2Fe(CO) 14Fe 3(CO) 12504823Mn-Fe,281.5Fe-Fe,281.5Mn Fe MnFeFeFeOs 3H 2(CO) 104642Os-Os,281.5Os[Mo 3(μ 3-O)(μ 2-O) 3F 9)] 5-Re 3(μ 2-Cl) 3(CH2SiMe 3) 6423669Os=Os,268.0Mo=Mo,250.2Re≡Re,238.7OsMoReMoOsMoReRe
Tetranuclear compoundsIr 4 (CO) 12 Re 4 (CO) 162-Os 4 (CO) 16g=4*9+12*2=60 g=4*7+16*2+2=62 g=4*9+16*2=64b=1/2(18*4-60)=6 b=1/2(18*4-62)=5 b=1/2(18*4-64)=4Pentanuclear compoundsHexanuclear compounds
Multi-nuclear (N>6) compounds6.5 Carbon clusters and nanotubes1. FullerencesC 50 Cl 10
2. Carbon nanotubes200 nm 1µm6.6 Hydrogen Bonding1.85Å0.95Å* Hydrogen bonding in DNA