Course program overview
Course program overview
Course program overview
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
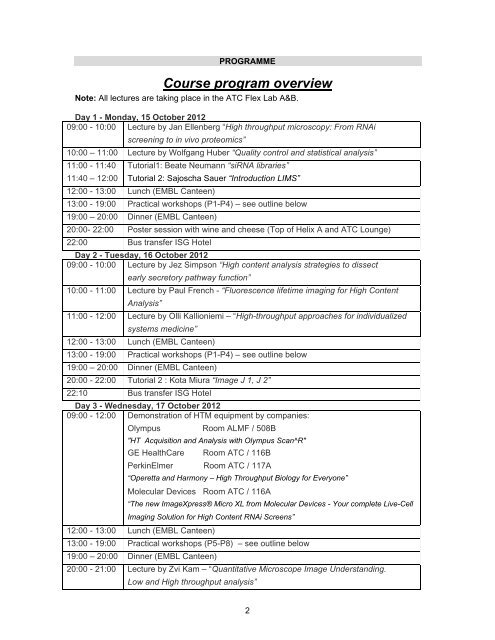
PROGRAMME<br />
<strong>Course</strong> <strong>program</strong> <strong>overview</strong><br />
Note: All lectures are taking place in the ATC Flex Lab A&B.<br />
Day 1 - Monday, 15 October 2012<br />
09:00 - 10:00 Lecture by Jan Ellenberg “High throughput microscopy: From RNAi<br />
screening to in vivo proteomics”<br />
10:00 – 11:00 Lecture by Wolfgang Huber “Quality control and statistical analysis”<br />
11:00 - 11:40<br />
11:40 – 12:00<br />
Tutorial1: Beate Neumann “siRNA libraries”<br />
Tutorial 2: Sajoscha Sauer “Introduction LIMS”<br />
12:00 - 13:00 Lunch (EMBL Canteen)<br />
13:00 - 19:00 Practical workshops (P1-P4) – see outline below<br />
19:00 – 20:00 Dinner (EMBL Canteen)<br />
20:00- 22:00 Poster session with wine and cheese (Top of Helix A and ATC Lounge)<br />
22:00 Bus transfer ISG Hotel<br />
Day 2 - Tuesday, 16 October 2012<br />
09:00 - 10:00 Lecture by Jez Simpson “High content analysis strategies to dissect<br />
early secretory pathway function”<br />
10:00 - 11:00 Lecture by Paul French - “Fluorescence lifetime imaging for High Content<br />
Analysis”<br />
11:00 - 12:00 Lecture by Olli Kallioniemi – “High-throughput approaches for individualized<br />
systems medicine”<br />
12:00 - 13:00 Lunch (EMBL Canteen)<br />
13:00 - 19:00 Practical workshops (P1-P4) – see outline below<br />
19:00 – 20:00 Dinner (EMBL Canteen)<br />
20:00 - 22:00 Tutorial 2 : Kota Miura “Image J 1, J 2”<br />
22:10 Bus transfer ISG Hotel<br />
Day 3 - Wednesday, 17 October 2012<br />
09:00 - 12:00 Demonstration of HTM equipment by companies:<br />
Olympus Room ALMF / 508B<br />
"HT Acquisition and Analysis with Olympus Scan^R"<br />
GE HealthCare Room ATC / 116B<br />
PerkinElmer Room ATC / 117A<br />
“Operetta and Harmony – High Throughput Biology for Everyone”<br />
Molecular Devices Room ATC / 116A<br />
“The new ImageXpress® Micro XL from Molecular Devices - Your complete Live-Cell<br />
Imaging Solution for High Content RNAi Screens”<br />
12:00 - 13:00 Lunch (EMBL Canteen)<br />
13:00 - 19:00 Practical workshops (P5-P8) – see outline below<br />
19:00 – 20:00 Dinner (EMBL Canteen)<br />
20:00 - 21:00 Lecture by Zvi Kam – “Quantitative Microscope Image Understanding.<br />
Low and High throughput analysis”<br />
2
21:10 Bus transfer ISG Hotel<br />
Day 4 - Thursday, 18 October 2012<br />
09:00 - 10:00 Lecture by Daniel Gehrlich “Supervised and unsupervised machine<br />
learning for image-based screening”<br />
10:00-11:00 Lecture by Seamus Holden “Towards high-throughput super-resolution<br />
microscopy in bacteria”<br />
11:00 - 12:00 Lecture by Thomas Walter -"From images to gene clusters: computational<br />
approaches for phenotypic profiling"<br />
12:00 - 13:00 Lunch (EMBL Canteen)<br />
13:00 - 19:00 Practical workshops (P5-P8) – see outline below<br />
19:00 – 20:00 Dinner (EMBL Canteen)<br />
20:00 Bus transfer to ISG or downtown Heidelberg<br />
Day 5 - Friday, 19 October 2012<br />
09:00 - 10:00 Lecture by Claudia Lukas “Exploring the mammalian DNA damage<br />
response using high content siRNA screens: three short stories based<br />
on one lucky readout”<br />
10:00 -11:00 Lecture by Michael Boutros "Mapping of genetic interaction networks<br />
by RNAi and high-throughput imaging"<br />
11:00 - 12:00 Lecture by Malte Wachsmuth "High throughput FCS: parallelization<br />
and automation"<br />
12:00 - 13:00 Lunch (EMBL Canteen)<br />
13:00 - 19:00 Practical workshops (P9-P12) – see outline below<br />
19.00 - 20.00 Bus transfer via ISG to downtown Heidelberg<br />
20:00 Dinner downtown Heidelberg (Restaurant Zum Nepomuk)<br />
Individual transport to ISG Hotel<br />
Day 6 - Saturday, 20 October 2012<br />
09:00 - 10:00 Lecture by Rainer Pepperkok “High throughput microscopy approaches<br />
to disease mechanism”<br />
10:00 - 13:00 Practical workshops (P9-P12) – see outline below<br />
13:00 - 14:00 Lunch (EMBL Canteen)<br />
14:00 - 17:00 Practical workshops (P9-P12) – see outline below<br />
17:00 -18:00 Summary of the course and discussion<br />
18:10 Bus transfer to ISG Hotel<br />
3
Monday and Tuesday P1-P4<br />
Practical work<br />
P1: Coating multiwall plates ready for reverse transfection. H. Erfle and F. Neumann<br />
P2: Sample preparation. J. Bulkescher and B. Koch<br />
P3: Assay design: C. Conrad, J. Ellenberg, R. Pepperkok and J. Simpson<br />
P4: Concepts and tools for quality control. B. Fischer, J. Barry, F. Klein, Ch. Tischer<br />
Wednesday and Thursday P5-P8<br />
P5: Time-lapse acquisition of mitotic phenotypes. F. Neumann and M. Rommeswinkel<br />
(Olympus)<br />
P6: Automated FCS- data acquisition C. Conrad and M. Wachsmuth<br />
P7: Inner nuclear membrane targeting. A. Boni and Ch. Tischer<br />
P8: Microtubuli secondary screening. J. Bulkescher, F. Sieckmann and Jan Kaluza (Leica)<br />
Friday and Saturday P9-P12<br />
P9: Image analysis of P5 (time-lapse data). Ch. Sommer and T. Walter<br />
P10: Image and FCS data analysis of P6. C. Conrad and M. Wachsmuth<br />
P11: Image analysis of P7. A. Boni and K. Miura<br />
P12: Large scale analysis and Bioinformatics. B. Fischer, W. Huber, J. Barry and F.Klein<br />
Outline of the practical work<br />
Monday and Tuesday<br />
P1 Coating 96 well plates ready for transfection<br />
(ALMF screening lab / building 13, room 507)<br />
Staff: Beate Neumann, Holger Erfle<br />
Work:<br />
1. Preparation of transfection solutions with the Liquidator.<br />
2. Coating of 96 well plate<br />
3. Discussion on plate layout, QC, long-term storage.<br />
4. Inspection of coated plates.<br />
P2 Sample preparation<br />
(ALMF cell culture lab / building 13, room 505)<br />
Staff: Jutta Bulkescher, Birgit Koch<br />
Work:<br />
1. Plating cells on transfection ready 96 well plates using the cell seeding<br />
device.<br />
2. Discussion on live cell imaging requirements<br />
4
P3 Assay design<br />
(ATC / Flex Lab)<br />
Staff: C. Conrad, J. Ellenberg, R. Pepperkok and J. Simpson<br />
Blackboard Session:<br />
1. Examples of working assays, cell systems.<br />
2. Dynamic range, reproducibility, controls, specificity.<br />
3. Assay design and image analysis.<br />
4. Assays design and suitability for HCS microscopy.<br />
P4 Batch image analysis and quality control<br />
(ATC / Flex Lab B)<br />
Staff: C. Tischer<br />
Work:<br />
1. Batch image analysis using CellProfiler<br />
2. Data examination using CellProfilerAnalyst<br />
Wednesday and Thursday<br />
P5 Image acquisition time-lapse<br />
(ALMF building 13, room 508B)<br />
Staff: Beate Neumann, Matthias Rommeswinkel (Olympus)<br />
Work:<br />
1. Discussion of general set-up and functions of the microscope system.<br />
2. Optimising parameters (e.g. AF, illumination) for live cell image acquisition.<br />
3. Defining plate layout on ScanR system.<br />
4. Image acquisition by students.<br />
P6 Automated fluorescence correlation spectroscopy (FCS)<br />
(MitoSys microscopy lab / building 1D, room 4.52)<br />
Staff: Christian Conrad, Malte Wachsmuth<br />
Work:<br />
1. Introduction to manual fluorescence correlation spectroscopy (FCS)<br />
experiments<br />
2. Automated acquisition of FCS data in 96 well plates<br />
5
P7 Inner nuclear membrane targeting<br />
(ALMF / building 13, room 513)<br />
Staff: Andrea Boni, Christian Tischer<br />
Work:<br />
1. Introduction to an assay to image Inner Nuclear Membrane protein (INMP)<br />
targeting in living cells<br />
2. Setup of time-lapse microscopy experiment using confocal microscope<br />
combined with automated cell recognition and drug dispenser<br />
3. Acquisition of live cell data of INMP targeting<br />
P8 Image acquisition secondary screen<br />
(ALMF building 13, room 508F)<br />
Staff: Jutta Bulkescher, Volker Hilsenstein, Jan Kaluza, Frank Sieckmann<br />
Work:<br />
1. Automated high resolution image acquisition<br />
2. Micropilot: classifier training for automatic online classification<br />
Friday and Saturday<br />
P9 Image analysis of P5 (time-lapse data)<br />
(ATC, Flex Lab B)<br />
Staff: Christoph Sommer, Thomas Walter<br />
Work:<br />
1. Discussion of the general strategy and practical issues.<br />
2. Step-by-step analysis of time-lapse data (from P5): segmentation, feature<br />
extraction, classification (including annotation and training of a classifier)<br />
and error correction.<br />
3. Application of the workflow to selected wells and performance evaluation.<br />
4. Browsing of analysis results (prepared) and biological interpretation.<br />
P10 Image and FCS data analysis of P6 (Automated FCS)<br />
(ATC, Flex Lab A)<br />
Staff: Christian Conrad, Malte Wachsmuth<br />
Work:<br />
1. Introduction to the biophysical background of FCS and to fundamentals of<br />
data fitting<br />
2. Analysis of the data acquired during P6 (Automated FCS)<br />
6
P11 Imaging analysis of Practical 7<br />
(Cell Biology and Biophysics Unit, room 417)<br />
Staff: Kota Miura, Andrea Boni<br />
Work:<br />
1. Basic introduction to the image analysis software ImageJ<br />
2. Imaging analysis of INM targeting time lapse microscopy data using<br />
ImageJ<br />
3. Automation of imaging processing by Macro scripting<br />
P12 Large Scale Analysis and Modelling<br />
(Room 6.51A Friday / 518 A+B Saturday)<br />
Staff: Joseph Barry, Felix Klein, Bernd Fischer<br />
Work:<br />
1. Quality control of high-throughput screening data using the<br />
R/bioconductor-package cell HTS2<br />
2. Plate normalization methods, correction of spatial plate effects,<br />
and scoring<br />
3. Downstream bioinformatics analysis (gene set enrichment<br />
analysis) and interpretation<br />
7