Review the symposium abstracts (3.19MB PDF) - College of Science
Review the symposium abstracts (3.19MB PDF) - College of Science
Review the symposium abstracts (3.19MB PDF) - College of Science
- No tags were found...
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
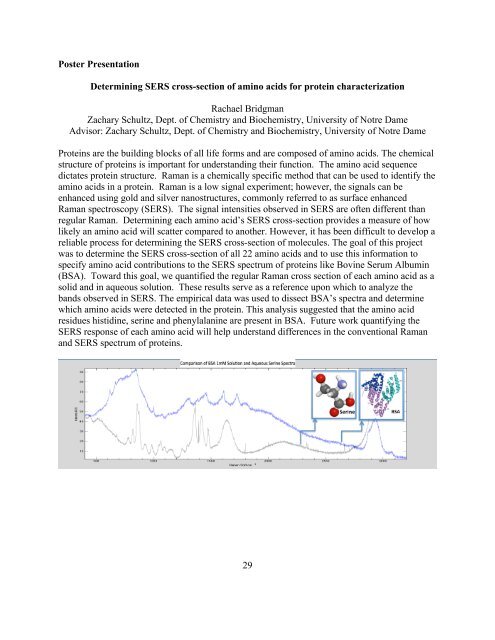
Poster PresentationDetermining SERS cross-section <strong>of</strong> amino acids for protein characterizationRachael BridgmanZachary Schultz, Dept. <strong>of</strong> Chemistry and Biochemistry, University <strong>of</strong> Notre DameAdvisor: Zachary Schultz, Dept. <strong>of</strong> Chemistry and Biochemistry, University <strong>of</strong> Notre DameProteins are <strong>the</strong> building blocks <strong>of</strong> all life forms and are composed <strong>of</strong> amino acids. The chemicalstructure <strong>of</strong> proteins is important for understanding <strong>the</strong>ir function. The amino acid sequencedictates protein structure. Raman is a chemically specific method that can be used to identify <strong>the</strong>amino acids in a protein. Raman is a low signal experiment; however, <strong>the</strong> signals can beenhanced using gold and silver nanostructures, commonly referred to as surface enhancedRaman spectroscopy (SERS). The signal intensities observed in SERS are <strong>of</strong>ten different thanregular Raman. Determining each amino acid’s SERS cross-section provides a measure <strong>of</strong> howlikely an amino acid will scatter compared to ano<strong>the</strong>r. However, it has been difficult to develop areliable process for determining <strong>the</strong> SERS cross-section <strong>of</strong> molecules. The goal <strong>of</strong> this projectwas to determine <strong>the</strong> SERS cross-section <strong>of</strong> all 22 amino acids and to use this information tospecify amino acid contributions to <strong>the</strong> SERS spectrum <strong>of</strong> proteins like Bovine Serum Albumin(BSA). Toward this goal, we quantified <strong>the</strong> regular Raman cross section <strong>of</strong> each amino acid as asolid and in aqueous solution. These results serve as a reference upon which to analyze <strong>the</strong>bands observed in SERS. The empirical data was used to dissect BSA’s spectra and determinewhich amino acids were detected in <strong>the</strong> protein. This analysis suggested that <strong>the</strong> amino acidresidues histidine, serine and phenylalanine are present in BSA. Future work quantifying <strong>the</strong>SERS response <strong>of</strong> each amino acid will help understand differences in <strong>the</strong> conventional Ramanand SERS spectrum <strong>of</strong> proteins.29