Hyperspectral Vegetation Indices and Their Relationships with ...
Hyperspectral Vegetation Indices and Their Relationships with ...
Hyperspectral Vegetation Indices and Their Relationships with ...
- No tags were found...
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
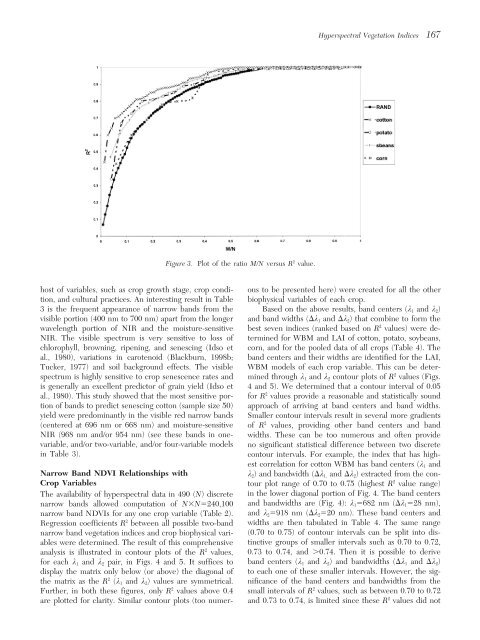
<strong>Hyperspectral</strong> <strong>Vegetation</strong> <strong>Indices</strong> 167Figure 3. Plot of the ratio M/N versus R 2 value.host of variables, such as crop growth stage, crop condi- ous to be presented here) were created for all the othertion, <strong>and</strong> cultural practices. An interesting result in Table biophysical variables of each crop.3 is the frequent appearance of narrow b<strong>and</strong>s from the Based on the above results, b<strong>and</strong> centers (k 1 <strong>and</strong> k 2 )visible portion (400 nm to 700 nm) apart from the longer <strong>and</strong> b<strong>and</strong> widths (k 1 <strong>and</strong> k 2 ) that combine to form thewavelength portion of NIR <strong>and</strong> the moisture-sensitive best seven indices (ranked based on R 2 values) were determinedNIR. The visible spectrum is very sensitive to loss offor WBM <strong>and</strong> LAI of cotton, potato, soybeans,chlorophyll, browning, ripening, <strong>and</strong> senescing (Idso et corn, <strong>and</strong> for the pooled data of all crops (Table 4). Theal., 1980), variations in carotenoid (Blackburn, 1998b; b<strong>and</strong> centers <strong>and</strong> their widths are identified for the LAI,Tucker, 1977) <strong>and</strong> soil background effects. The visible WBM models of each crop variable. This can be determinedspectrum is highly sensitive to crop senescence rates <strong>and</strong>through k 1 <strong>and</strong> k 2 contour plots of R 2 values (Figs.is generally an excellent predictor of grain yield (Idso et 4 <strong>and</strong> 5). We determined that a contour interval of 0.05al., 1980). This study showed that the most sensitive portionfor R 2 values provide a reasonable <strong>and</strong> statistically soundof b<strong>and</strong>s to predict senescing cotton (sample size 50) approach of arriving at b<strong>and</strong> centers <strong>and</strong> b<strong>and</strong> widths.yield were predominantly in the visible red narrow b<strong>and</strong>s Smaller contour intervals result in several more gradients(centered at 696 nm or 668 nm) <strong>and</strong> moisture-sensitive of R 2 values, providing other b<strong>and</strong> centers <strong>and</strong> b<strong>and</strong>NIR (968 nm <strong>and</strong>/or 954 nm) (see these b<strong>and</strong>s in one- widths. These can be too numerous <strong>and</strong> often providevariable, <strong>and</strong>/or two-variable, <strong>and</strong>/or four-variable models no significant statistical difference between two discretein Table 3).contour intervals. For example, the index that has highestcorrelation for cotton WBM has b<strong>and</strong> centers (k 1 <strong>and</strong>Narrow B<strong>and</strong> NDVI <strong>Relationships</strong> <strong>with</strong>k 2 ) <strong>and</strong> b<strong>and</strong>width (k 1, <strong>and</strong> k 2 ) extracted from the con-Crop Variables tour plot range of 0.70 to 0.75 (highest R 2 value range)The availability of hyperspectral data in 490 (N) discrete in the lower diagonal portion of Fig. 4. The b<strong>and</strong> centersnarrow b<strong>and</strong>s allowed computation of NN240,100 <strong>and</strong> b<strong>and</strong>widths are (Fig. 4): k 1 682 nm (k 1 28 nm),narrow b<strong>and</strong> NDVIs for any one crop variable (Table 2). <strong>and</strong> k 2 918 nm (k 2 20 nm). These b<strong>and</strong> centers <strong>and</strong>Regression coefficients R 2 between all possible two-b<strong>and</strong> widths are then tabulated in Table 4. The same rangenarrow b<strong>and</strong> vegetation indices <strong>and</strong> crop biophysical varitinctive(0.70 to 0.75) of contour intervals can be split into dis-ables were determined. The result of this comprehensivegroups of smaller intervals such as 0.70 to 0.72,analysis is illustrated in contour plots of the R 2 values, 0.73 to 0.74, <strong>and</strong> 0.74. Then it is possible to derivefor each k 1 <strong>and</strong> k 2 pair, in Figs. 4 <strong>and</strong> 5. It suffices to b<strong>and</strong> centers (k 1 <strong>and</strong> k 2 ) <strong>and</strong> b<strong>and</strong>widths (k 1 <strong>and</strong> k 2 )display the matrix only below (or above) the diagonal of to each one of these smaller intervals. However, the sig-the matrix as the R 2 (k 1 <strong>and</strong> k 2 ) values are symmetrical. nificance of the b<strong>and</strong> centers <strong>and</strong> b<strong>and</strong>widths from theFurther, in both these figures, only R 2 values above 0.4 small intervals of R 2 values, such as between 0.70 to 0.72are plotted for clarity. Similar contour plots (too numer- <strong>and</strong> 0.73 to 0.74, is limited since these R 2 values did not