EMBL-EBI Annual Scientific Report 2012
EMBL-EBI Annual Scientific Report 2012
EMBL-EBI Annual Scientific Report 2012
- No tags were found...
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
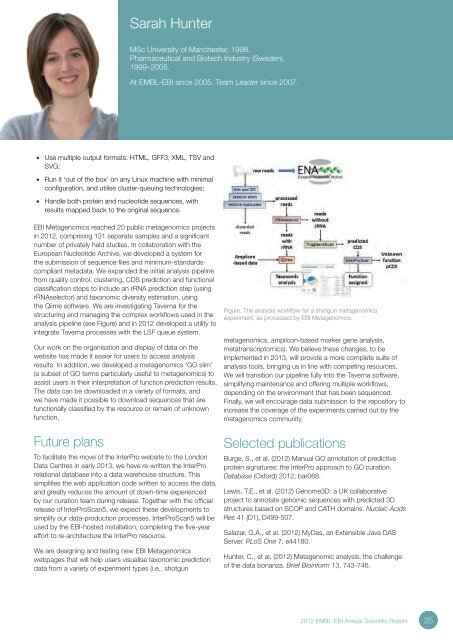
Sarah HunterMSc University of Manchester, 1998.Pharmaceutical and Biotech Industry (Sweden),1999–2005.At <strong>EMBL</strong>-<strong>EBI</strong> since 2005. Team Leader since 2007.• Use multiple output formats: HTML, GFF3, XML, TSV andSVG;• Run it ‘out of the box’ on any Linux machine with minimalconfiguration, and utilise cluster-queuing technologies;• Handle both protein and nucleotide sequences, withresults mapped back to the original sequence.<strong>EBI</strong> Metagenomics reached 20 public metagenomics projectsin <strong>2012</strong>, comprising 131 separate samples and a significantnumber of privately held studies. In collaboration with theEuropean Nucleotide Archive, we developed a system forthe submission of sequence files and minimum-standardscompliantmetadata. We expanded the initial analysis pipelinefrom quality control, clustering, CDS prediction and functionalclassification steps to include an rRNA prediction step (usingrRNAselector) and taxonomic diversity estimation, usingthe Qiime software. We are investigating Taverna for thestructuring and managing the complex workflows used in theanalysis pipeline (see Figure) and in <strong>2012</strong> developed a utility tointegrate Taverna processes with the LSF queue system.Our work on the organisation and display of data on thewebsite has made it easier for users to access analysisresults. In addition, we developed a metagenomics ‘GO slim’(a subset of GO terms particularly useful to metagenomics) toassist users in their interpretation of function prediction results.The data can be downloaded in a variety of formats, andwe have made it possible to download sequences that arefunctionally classified by the resource or remain of unknownfunction.Future plansTo facilitate the move of the InterPro website to the LondonData Centres in early 2013, we have re-written the InterProrelational database into a data warehouse structure. Thissimplifies the web application code written to access the data,and greatly reduces the amount of down-time experiencedby our curation team during release. Together with the officialrelease of InterProScan5, we expect these developments tosimplify our data-production processes. InterProScan5 will beused by the <strong>EBI</strong>-hosted installation, completing the five-yeareffort to re-architecture the InterPro resource.We are designing and testing new <strong>EBI</strong> Metagenomicswebpages that will help users visualise taxonomic predictiondata from a variety of experiment types (i.e., shotgunFigure. The analysis workflow for a shotgun metagenomicsexperiment, as processed by <strong>EBI</strong> Metagenomics.metagenomics, amplicon-based marker gene analysis,metatranscriptomics). We believe these changes, to beimplemented in 2013, will provide a more complete suite ofanalysis tools, bringing us in line with competing resources.We will transition our pipeline fully into the Taverna software,simplifying maintenance and offering multiple workflows,depending on the environment that has been sequenced.Finally, we will encourage data submission to the repository toincrease the coverage of the experiments carried out by themetagenomics community.Selected publicationsBurge, S., et al. (<strong>2012</strong>) Manual GO annotation of predictiveprotein signatures: the InterPro approach to GO curation.Database (Oxford) <strong>2012</strong>, bar068.Lewis, T.E., et al. (<strong>2012</strong>) Genome3D: a UK collaborativeproject to annotate genomic sequences with predicted 3Dstructures based on SCOP and CATH domains. Nucleic AcidsRes 41 (D1), D499-507.Salazar, G.A., et al. (<strong>2012</strong>) MyDas, an Extensible Java DASServer. PLoS One 7, e44180.Hunter, C., et al. (<strong>2012</strong>) Metagenomic analysis: the challengeof the data bonanza. Brief Bioinform 13, 743-746.<strong>2012</strong> <strong>EMBL</strong>-<strong>EBI</strong> <strong>Annual</strong> <strong>Scientific</strong> <strong>Report</strong>35