RNA Splicing: Removal of Introns from Primary ... - EURASNET
RNA Splicing: Removal of Introns from Primary ... - EURASNET
RNA Splicing: Removal of Introns from Primary ... - EURASNET
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
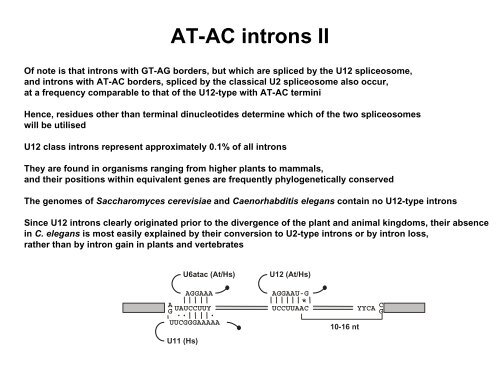
AT-AC introns IIOf note is that introns with GT-AG borders, but which are spliced by the U12 spliceosome,and introns with AT-AC borders, spliced by the classical U2 spliceosome also occur,at a frequency comparable to that <strong>of</strong> the U12-type with AT-AC terminiHence, residues other than terminal dinucleotides determine which <strong>of</strong> the two spliceosomeswill be utilisedU12 class introns represent approximately 0.1% <strong>of</strong> all intronsThey are found in organisms ranging <strong>from</strong> higher plants to mammals,and their positions within equivalent genes are frequently phylogenetically conservedThe genomes <strong>of</strong> Saccharomyces cerevisiae and Caenorhabditis elegans contain no U12-type intronsSince U12 introns clearly originated prior to the divergence <strong>of</strong> the plant and animal kingdoms, their absencein C. elegans is most easily explained by their conversion to U2-type introns or by intron loss,rather than by intron gain in plants and vertebratesAGU6atac (At/Hs)AGGAAAUAUCCUUYUUCGGGAAAAAU11 (Hs)U12 (At/Hs)AGGAAU-GUCCUUAAC10-16 ntYYCACG