2008 Scientific Report
Create successful ePaper yourself
Turn your PDF publications into a flip-book with our unique Google optimized e-Paper software.
VARI | <strong>2008</strong><br />
Research Interests<br />
We are studying how cells accurately replicate their DNA, a process that begins at specific DNA sequences termed replication<br />
origins. Various genome-wide approaches have identified from 320 to 420 possible replication origins in budding yeast,<br />
the model organism we study. In G1 phase, each origin assembles approximately 40 polypeptides in a temporally defined<br />
order, culminating in the initiation of DNA replication at the G1/S phase boundary. The first stage of this process is called<br />
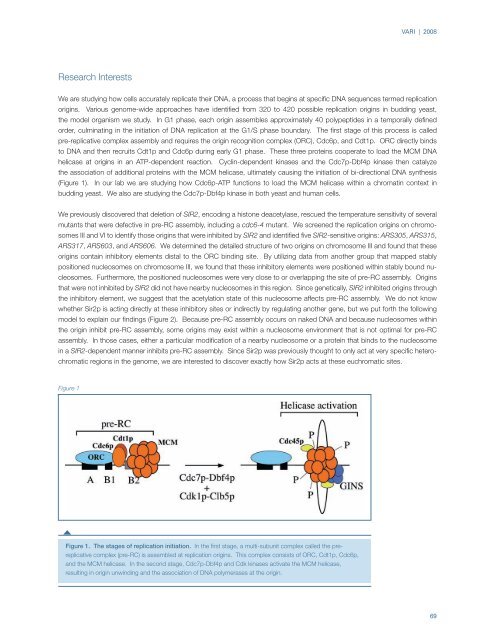
pre-replicative complex assembly and requires the origin recognition complex (ORC), Cdc6p, and Cdt1p. ORC directly binds<br />
to DNA and then recruits Cdt1p and Cdc6p during early G1 phase. These three proteins cooperate to load the MCM DNA<br />
helicase at origins in an ATP-dependent reaction. Cyclin-dependent kinases and the Cdc7p-Dbf4p kinase then catalyze<br />
the association of additional proteins with the MCM helicase, ultimately causing the initiation of bi-directional DNA synthesis<br />
(Figure 1). In our lab we are studying how Cdc6p-ATP functions to load the MCM helicase within a chromatin context in<br />
budding yeast. We also are studying the Cdc7p-Dbf4p kinase in both yeast and human cells.<br />
We previously discovered that deletion of SIR2, encoding a histone deacetylase, rescued the temperature sensitivity of several<br />
mutants that were defective in pre-RC assembly, including a cdc6-4 mutant. We screened the replication origins on chromosomes<br />
III and VI to identify those origins that were inhibited by SIR2 and identified five SIR2-sensitive origins: ARS305, ARS315,<br />
ARS317, ARS603, and ARS606. We determined the detailed structure of two origins on chromosome III and found that these<br />
origins contain inhibitory elements distal to the ORC binding site. By utilizing data from another group that mapped stably<br />
positioned nucleosomes on chromosome III, we found that these inhibitory elements were positioned within stably bound nucleosomes.<br />
Furthermore, the positioned nucleosomes were very close to or overlapping the site of pre-RC assembly. Origins<br />
that were not inhibited by SIR2 did not have nearby nucleosomes in this region. Since genetically, SIR2 inhibited origins through<br />
the inhibitory element, we suggest that the acetylation state of this nucleosome affects pre-RC assembly. We do not know<br />
whether Sir2p is acting directly at these inhibitory sites or indirectly by regulating another gene, but we put forth the following<br />
model to explain our findings (Figure 2). Because pre-RC assembly occurs on naked DNA and because nucleosomes within<br />
the origin inhibit pre-RC assembly, some origins may exist within a nucleosome environment that is not optimal for pre-RC<br />
assembly. In those cases, either a particular modification of a nearby nucleosome or a protein that binds to the nucleosome<br />
in a SIR2-dependent manner inhibits pre-RC assembly. Since Sir2p was previously thought to only act at very specific heterochromatic<br />
regions in the genome, we are interested to discover exactly how Sir2p acts at these euchromatic sites.<br />
Figure 1<br />
Figure 1. The stages of replication initiation. In the first stage, a multi-subunit complex called the prereplicative<br />
complex (pre-RC) is assembled at replication origins. This complex consists of ORC, Cdt1p, Cdc6p,<br />
and the MCM helicase. In the second stage, Cdc7p-Dbf4p and Cdk kinases activate the MCM helicase,<br />
resulting in origin unwinding and the association of DNA polymerases at the origin.<br />
69