You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
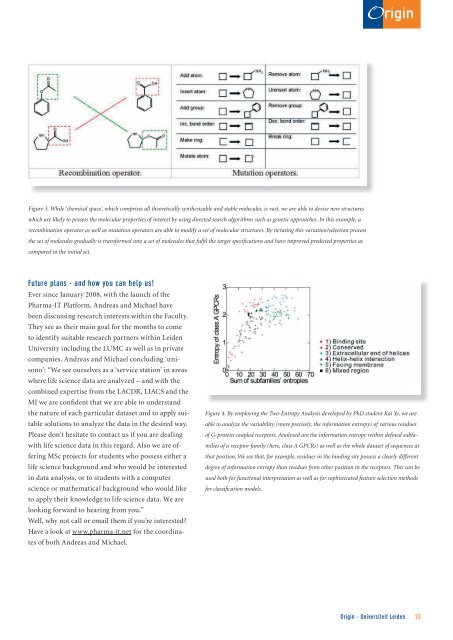
Figure 3. While ‘chemical space’, which comprises all theoretically synthesizable and stable molecules, is vast, we are able to devise new structures<br />
which are likely to possess the molecular properties of interest by using directed search algorithms such as genetic approaches. In this example, a<br />
recombination operator as well as mutation operators are able to modify a set of molecular structures. By iterating this variation/selection process<br />
the set of molecules gradually is transformed into a set of molecules that fulfil the target specifications and have improved predicted properties as<br />
compared to the initial set.<br />
Future plans - and how you can help us!<br />
Ever since January <strong>2008</strong>, with the launch of the<br />
Pharma-IT Platform, Andreas and Michael have<br />
been discussing research interests within the Faculty.<br />
They see as their main goal for the months to come<br />
to identify suitable research partners within Leiden<br />
University including the LUMC as well as in private<br />
companies. Andreas and Michael concluding ‘unisono’:<br />
“We see ourselves as a ‘service station’ in areas<br />
where life science data are analyzed – and with the<br />
combined expertise from the LACDR, LIACS and the<br />
MI we are confident that we are able to understand<br />
the nature of each particular dataset and to apply suitable<br />
solutions to analyze the data in the desired way.<br />
Please don’t hesitate to contact us if you are dealing<br />
with life science data in this regard. Also we are offering<br />
MSc projects for students who possess either a<br />
life science background and who would be interested<br />
in data analysis, or to students with a computer<br />
science or mathematical background who would like<br />
to apply their knowledge to life science data. We are<br />
looking forward to hearing from you.”<br />
Well, why not call or email them if you’re interested?<br />
Have a look at www.pharma-it.net for the coordinates<br />
of both Andreas and Michael.<br />
Figure 4. By employing the Two-Entropy Analysis developed by PhD student Kai Ye, we are<br />
able to analyze the variability (more precisely, the information entropy) of various residues<br />
of G-protein coupled receptors. Analyzed are the information entropy within defined subfamilies<br />
of a receptor family (here, class A GPCRs) as well as the whole dataset of sequences at<br />
that position. We see that, for example, residues in the binding site possess a clearly different<br />
degree of information entropy than residues from other position in the receptors. This can be<br />
used both for functional interpretation as well as for sophisticated feature selection methods<br />
for classification models.<br />
Origin - Universiteit Leiden 23