download/files/Proteomics brochure 7-08-04.pdf
download/files/Proteomics brochure 7-08-04.pdf
download/files/Proteomics brochure 7-08-04.pdf
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
Phosphorylation Site Mapping<br />
Identify sites of protein phosphorylation using chromatography columns that provide a high yield of<br />
phosphopeptides plus Data Dependent Neutral Loss MS 3<br />
scanning with the Finnigan LTQ to selectively<br />
perform MS 3<br />
on phosphopeptides.<br />
Relative Abundance<br />
100<br />
50<br />
100<br />
90<br />
80<br />
70<br />
60<br />
50<br />
40<br />
30<br />
20<br />
10<br />
20 .1 5<br />
22 .6 4<br />
27 .6 3<br />
33 .1 3<br />
28 .2 0<br />
33 .5 9<br />
35 .7 6<br />
37 .3 6<br />
39 .5 2<br />
41 .4 0<br />
MS<br />
MS 2<br />
MS 3<br />
41 .7 1<br />
19 .7 3<br />
0<br />
4. 55 9. 44<br />
15 .4 6 42 .7 6 49 .0 4 57 .3 4<br />
0 10 20 30 40 50 60<br />
100<br />
0<br />
476.27 706.<strong>08</strong><br />
688.29<br />
1031.78<br />
1058.62<br />
982.624<br />
747.31 1013.78<br />
503.37 632.39 876.39<br />
1105.42<br />
390.18<br />
844.91<br />
1375.65<br />
1490.75<br />
1361.58<br />
1619 .71<br />
1619.72<br />
80<br />
964.67<br />
60<br />
747.34<br />
1688.81<br />
632.38<br />
40 328.26<br />
503.42<br />
20<br />
603.30<br />
345.07<br />
827.52<br />
1<strong>08</strong>8.34<br />
859.53<br />
1105.56 1361 .71 1601.85<br />
1332 .52<br />
1233.69<br />
1491.61<br />
1070.57<br />
1199.39<br />
1574.61<br />
1817.71<br />
1800 .74<br />
Time (min) m/z<br />
0<br />
0 5 10 15 20 25 30 35 40 45 50 55 60 65 70 75 80 85 90 95 100<br />
Time (min)<br />
100<br />
50<br />
0<br />
0 10 20 30 40 50 60 70 80 90 100 110 120 130 140 150 160<br />
Time (min)<br />
100<br />
50<br />
0<br />
0 10 20 30 40 50<br />
100<br />
50<br />
0<br />
40 m M<br />
150 160<br />
0 10 20 30 40 50 60 70 80<br />
Time (min)<br />
90 100 110 120 130 140<br />
100<br />
50<br />
0<br />
0 10 20 30 40 50 60 70 80 90 100 110 120 130 140 150 160<br />
Time (min)<br />
100<br />
50<br />
80 m M<br />
0<br />
0 10 20 30 40 50 60 70 80 90 100 110 120 130 140 150 160<br />
Time (min)<br />
100<br />
50<br />
0<br />
0 10 20 30 40 50 60 70 80 90 100 110 120 130 140 150 160<br />
Time (min)<br />
100<br />
50<br />
0<br />
0 10 20 30 40 50 60 70 80 90 100 110 120 130 140 150 160<br />
Time (min)<br />
100<br />
50<br />
0<br />
0 10 20 30 40 50 60 70 80 90 100 110 120 130 140 150 160<br />
Time (min)<br />
100<br />
50<br />
0<br />
0 10 20 30 40 50 60 70 80 90 100 110 120 130 140 150 160<br />
Time (min)<br />
100<br />
50<br />
0<br />
0 10 20 30 40 50 60 70 80 90 100 110 120 130 140 150 160<br />
Time (min)<br />
100<br />
50<br />
(Shows<br />
neutral loss)<br />
0 m M<br />
10 m M<br />
20 m M<br />
60 m M<br />
100 m M<br />
120 m M<br />
150 m M<br />
200 m M<br />
400 m M<br />
800 m M<br />
0<br />
0 10 20 30 40 50 60 70 80 90 100 110 120 130 140 150 160<br />
Time (min)<br />
Relative Abundance<br />
40 0 60 0 80 0 1000 1200 1400 1600 1800 2000<br />
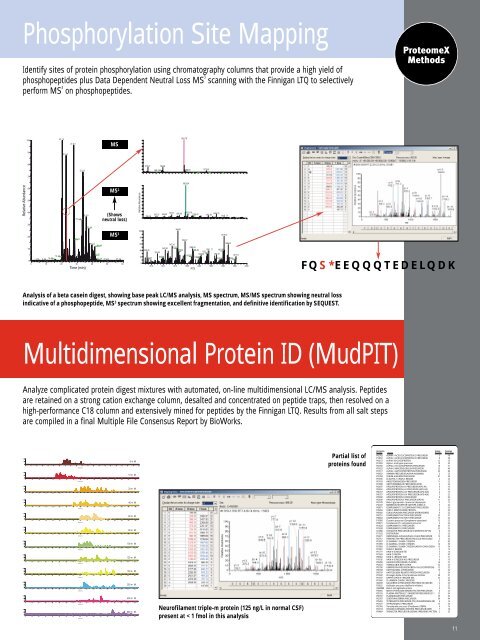
Analysis of a beta casein digest, showing base peak LC/MS analysis, MS spectrum, MS/MS spectrum showing neutral loss<br />
indicative of a phosphopeptide, MS 3 spectrum showing excellent fragmentation, and definitive identification by SEQUEST.<br />
FQS*EEQQQTEDELQDK<br />
Multidimensional Protein ID (MudPIT)<br />
Analyze complicated protein digest mixtures with automated, on-line multidimensional LC/MS analysis. Peptides<br />
are retained on a strong cation exchange column, desalted and concentrated on peptide traps, then resolved on a<br />
high-performance C18 column and extensively mined for peptides by the Finnigan LTQ. Results from all salt steps<br />
are compiled in a final Multiple File Consensus Report by BioWorks.<br />
Neurofilament triple-m protein (125 ng/L in normal CSF)<br />
present at < 1 fmol in this analysis<br />
Partial list of<br />
proteins found<br />
ProteomeX<br />
Methods<br />
Accession Unique Coverage<br />
Number Protein Peptides Percentage<br />
P02763 ALPHA-1-ACID GLYCOPROTEIN 2 PRECURSOR 5 18<br />
P19652 ALPHA-1-ACID GLYCOPROTEIN 2 PRECURSOR 8 28<br />
P04217 ALPHA-1B-GLYCOPROTEIN 12 25<br />
P01009 Alpha-1-antitrypsin precursor 10 26<br />
P02765 ALPHA-2-HS-GLYCOPROTEIN PRECURSOR 10 31<br />
P01023 ALPHA-2-MACROGLOBULIN PRECURSOR 35 48<br />
P01011 ALPHA-1-ANTICHYMOTRYPSIN PRECURSOR 25 84<br />
P43652 AFAMIN PRECURSOR (ALPHA-ALBUMIN) 3 25<br />
P02768 SERUM ALBUMIN PRECURSOR 67 58<br />
P01876 IG ALPHA-1 CHAIN C REGION 2 25<br />
P01019 ANGIOTENSINOGEN PRECURSOR 11 25<br />
P010<strong>08</strong> ANTITHROMBIN-III PRECURSOR (ATIII) 3 24<br />
P02647 APOLIPOPROTEIN A-I PRECURSOR (APO-AI) 6 16<br />
P02652 APOLIPOPROTEIN A-II PRECURSOR (APO-AII) 5 20<br />
P06727 APOLIPOPROTEIN A-IV PRECURSOR (APO-AIV) 4 15<br />
P06727 APOLIPOPROTEIN A-IV PRECURSOR (APO-AIV) 7 25<br />
P05090 APOLIPOPROTEIN D PRECURSOR 8 49<br />
P02649 APOLIPOPROTEIN E PRECURSOR (APO-E) 11 32<br />
P02749 Beta-2-glycoprotein I precursor (Apolipopro 5 12<br />
P32247 BOMBESIN RECEPTOR SUBTYPE-3 (BRS-3) 5 24<br />
P09871 COMPLEMENT C1S COMPONENT PRECURSOR 2 35<br />
P49454 CENP-F KINETOCHORE PROTEIN 3 45<br />
P00450 CERULOPLASMIN PRECURSOR (FERROXIDASE) 4 25<br />
P00751 COMPLEMENT FACTOR B PRECURSOR 23 37<br />
P<strong>08</strong>603 COMPLEMENT FACTOR H PRECURSOR 15 42<br />
P10909 Clusterin precursor (Complement-associated 15 25<br />
P09871 Complement C1s component precursor 2 24<br />
P01024 COMPLEMENT C3 PRECURSOR 38 37<br />
P01028 COMPLEMENT C4 PRECURSOR 32 23<br />
Q12860 CONTACTIN PRECURSOR (GLYCOPROTEIN GP135) 6 16<br />
P11532 DYSTROPHIN 5 25<br />
P02671 FIBRINOGEN ALPHA/ALPHA-E CHAIN PRECURSOR 13 34<br />
P02751 FIBRONECTIN PRECURSOR (FN) (COLD-INSOLUBLE 3 24<br />
P01857 IG GAMMA-1 CHAIN C REGION 21 60<br />
P01859 IG GAMMA-2 CHAIN C REGION 14 30<br />
P01860 IG GAMMA-3 CHAIN C REGION (HEAVY CHAIN DISEA 13 34<br />
P01861 CHAIN C REGION 3 15<br />
P01777 HAIN V-III REGION TEI 3 14<br />
P01834 HAIN C REGION 3 50<br />
P80362 HAIN V-I REGION WAT 7 21<br />
P18136 HAIN V-III REGION HIC PRECURSOR 6 16<br />
P06396 GELSOLIN PRECURSOR, PLASMA 16 27<br />
P02023 HEMOGLOBIN BETA CHAIN 32 23<br />
P02790 HEMOPEXIN PRECURSOR (BETA-1B-GLYCOPROTEIN) 12 25<br />
P00738 HAPTOGLOBIN-2 PRECURSOR 15 25<br />
P00739 HAPTOGLOBIN-RELATED PROTEIN PRECURSOR 2 24<br />
P01042 Kininogen Alpha-2-thiol proteinase inhibitor 38 37<br />
P01597 KAPPA CHAIN V-I REGION DEE 32 23<br />
P01842 IG LAMBDA CHAIN C REGIONS 3 50<br />
Q92876 KALLIKREIN 6 PRECURSOR (PROTEASE M) (NEURO 2 23<br />
P29622 Kallistatin precursor (Kallikrein inhibitor) 2 25<br />
Q9UM88 Beta 2-microglobulin protein 9 70<br />
P36955 MENT EPITHELIUM-DERIVED FACTOR PRECURSOR 6 23<br />
P05155 PLASMA PROTEASE C1 INHIBITOR PRECURSOR (C1 I 13 34<br />
P00747 PLASMINOGEN PRECURSOR 33 44<br />
P02787 SEROTRANSFERRIN PRECURSOR 34 44<br />
P05452 TETRANECTIN PRECURSOR (TN) (PLASMINOGEN-KRI 6 16<br />
P07477 TRYPSINOGEN I PRECURSOR 15 33<br />
P02766 Transthyretin precursor (Prealbumin) (TBPA) 6 16<br />
P02774 VITAMIN D-BINDING PROTEIN PRECURSOR (DBP) 7 18<br />
P04004 TRONECTIN PRECURSOR (SERUM SPREADING FACTOR) 2 35<br />
11