Mutations in the mitochondrial thioredoxin reductase gene TXNRD2 ...
Mutations in the mitochondrial thioredoxin reductase gene TXNRD2 ...
Mutations in the mitochondrial thioredoxin reductase gene TXNRD2 ...
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
1124<br />
Confocal microscopy was performed with a Leica DM IRBE microscope<br />
(500 nm excitation and 550 nm emission), a Leica TCS SP2<br />
scanner, and Leica Confocal software (Leica Microsystems GmbH,<br />
Wetzlar, Germany). Pictures were processed by Adobe Photoshop<br />
CS3. Process<strong>in</strong>g <strong>in</strong>cluded <strong>the</strong> removal of unspecific background and<br />
addition of false colours (blue for DAPI, green for Alexa488, and red<br />
for Alexa568) as well as <strong>the</strong> assembly of an overlay of a representative<br />
optical section from each cell. Scale bars <strong>in</strong>cluded represent 10 mm.<br />
Treatment of mouse embryonic fibroblasts<br />
with L-buthion<strong>in</strong>e sulfoxim<strong>in</strong>e and<br />
determ<strong>in</strong>ation of cell viability<br />
Cells were treated with <strong>in</strong>creas<strong>in</strong>g concentrations of L-buthion<strong>in</strong>e sulfoxim<strong>in</strong>e<br />
(BSO; Sigma-Aldrich). The cell number was determ<strong>in</strong>ed 48 h<br />
later by trypan blue exclusion as described previously. 18<br />
Transmission electron microscopy<br />
Cells were harvested and fixed <strong>in</strong> 2.5% glutaraldehyde <strong>in</strong> 0.1 M sodium<br />
cacodylate buffer (pH 7.4; Electron Microscopy Sciences, Hatfield, PA,<br />
USA) and embedded <strong>in</strong> epoxy res<strong>in</strong> (epon 812; Electron Microscopy<br />
Sciences). Ultrath<strong>in</strong> sections were exam<strong>in</strong>ed with an EM 10 CR transmission<br />
electron microscope (Carl Zeiss, Inc.). For image acquisition, a<br />
MegaView III camera system (Olympus) was used.<br />
Statistical analyses<br />
Statistical analysis of patients and controls was performed with <strong>the</strong><br />
STATA SE 8.0 Statistics/Data analysis package (StataCorp LP, TX,<br />
USA). Genotypes were compared between patients and controls<br />
with two-sided x 2 test. Statistical analysis of <strong>the</strong> cellular studies was<br />
performed us<strong>in</strong>g SigmaStat 3.1 (Systat Software GmbH, Erkrath,<br />
Germany). With<strong>in</strong> each panel, wt and knockout cells express<strong>in</strong>g<br />
empty vector (mock), wt Txnrd2, and <strong>the</strong> different mutants were compared<br />
us<strong>in</strong>g all pairwise multiple comparison procedures (Holm–Sidak<br />
method). P-values ,0.05 were considered as significant.<br />
Homology modell<strong>in</strong>g<br />
We modelled <strong>the</strong> structure of human <strong>TXNRD2</strong> <strong>in</strong>clud<strong>in</strong>g FAD and<br />
NADPH accord<strong>in</strong>g to <strong>the</strong> crystal structure of human TXNRD1<br />
(monomers C and D, 22) 24 and mur<strong>in</strong>e Txnrd2, 25 us<strong>in</strong>g <strong>the</strong> Swiss-<br />
Model automated comparative prote<strong>in</strong> modell<strong>in</strong>g server (http://<br />
swissmodel.expasy.org/).<br />
Results<br />
Study populations<br />
To identify disease-associated <strong>TXNRD2</strong> mutations, we studied a<br />
cohort of 227 DCM patients and 683 <strong>in</strong>dividuals from a <strong>gene</strong>ral<br />
population sample. The basel<strong>in</strong>e characteristics of patients (n ¼<br />
227) and <strong>the</strong> <strong>gene</strong>ral population sample (n ¼ 683) are shown <strong>in</strong><br />
Table 1. The mean (+SD) ejection fraction <strong>in</strong> DCM patients was<br />
26.9 + 9.5 vs. 65.5 + 11.9% <strong>in</strong> <strong>the</strong> <strong>gene</strong>ral population sample.<br />
Sequenc<strong>in</strong>g of all exons and <strong>the</strong> SECIS region of <strong>TXNRD2</strong> <strong>in</strong> a<br />
subset of <strong>the</strong> DCM patients (n ¼ 96) yielded seven synonymous<br />
and seven non-synonymous <strong>TXNRD2</strong> variants (Table 2). Genotype<br />
distributions of non-synonymous variants <strong>in</strong> <strong>the</strong> DCM patients and<br />
<strong>the</strong> <strong>gene</strong>ral population sample were not significantly different<br />
(Table 3).<br />
D. Sibb<strong>in</strong>g et al.<br />
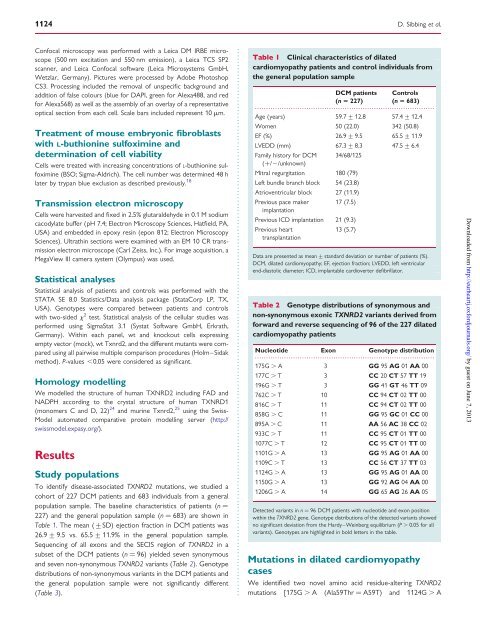
Table 1 Cl<strong>in</strong>ical characteristics of dilated<br />
cardiomyopathy patients and control <strong>in</strong>dividuals from<br />
<strong>the</strong> <strong>gene</strong>ral population sample<br />
DCM patients Controls<br />
(n 5 227) (n 5 683)<br />
................................................................................<br />
Age (years) 59.7 + 12.8 57.4 + 12.4<br />
Women 50 (22.0) 342 (50.8)<br />
EF (%) 26.9 + 9.5 65.5 + 11.9<br />
LVEDD (mm) 67.3 + 8.3 47.5 + 6.4<br />
Family history for DCM<br />
(+/2/unknown)<br />
34/68/125<br />
Mitral regurgitation 180 (79)<br />
Left bundle branch block 54 (23.8)<br />
Atrioventricular block 27 (11.9)<br />
Previous pace maker<br />
implantation<br />
17 (7.5)<br />
Previous ICD implantation 21 (9.3)<br />
Previous heart<br />
transplantation<br />
13 (5.7)<br />
Data are presented as mean + standard deviation or number of patients (%).<br />
DCM, dilated cardiomyopathy; EF, ejection fraction; LVEDD, left ventricular<br />
end-diastolic diameter; ICD, implantable cardioverter defibrillator.<br />
Table 2 Genotype distributions of synonymous and<br />
non-synonymous exonic <strong>TXNRD2</strong> variants derived from<br />
forward and reverse sequenc<strong>in</strong>g of 96 of <strong>the</strong> 227 dilated<br />
cardiomyopathy patients<br />
Nucleotide Exon Genotype distribution<br />
................................................................................<br />
175G . A 3 GG 95 AG 01 AA 00<br />
177C . T 3 CC 20 CT 57 TT 19<br />
196G . T 3 GG 41 GT 46 TT 09<br />
762C . T 10 CC 94 CT 02 TT 00<br />
816C . T 11 CC 94 CT 02 TT 00<br />
858G . C 11 GG 95 GC 01 CC 00<br />
895A . C 11 AA 56 AC 38 CC 02<br />
933C . T 11 CC 95 CT 01 TT 00<br />
1077C . T 12 CC 95 CT 01 TT 00<br />
1101G . A 13 GG 95 AG 01 AA 00<br />
1109C . T 13 CC 56 CT 37 TT 03<br />
1124G . A 13 GG 95 AG 01 AA 00<br />
1150G . A 13 GG 92 AG 04 AA 00<br />
1206G . A 14 GG 65 AG 26 AA 05<br />
Detected variants <strong>in</strong> n ¼ 96 DCM patients with nucleotide and exon position<br />
with<strong>in</strong> <strong>the</strong> <strong>TXNRD2</strong> <strong>gene</strong>. Genotype distributions of <strong>the</strong> detected variants showed<br />
no significant deviation from <strong>the</strong> Hardy–We<strong>in</strong>berg equilibrium (P . 0.05 for all<br />
variants). Genotypes are highlighted <strong>in</strong> bold letters <strong>in</strong> <strong>the</strong> table.<br />
<strong>Mutations</strong> <strong>in</strong> dilated cardiomyopathy<br />
cases<br />
We identified two novel am<strong>in</strong>o acid residue-alter<strong>in</strong>g <strong>TXNRD2</strong><br />
mutations [175G . A (Ala59Thr ¼ A59T) and 1124G . A<br />
Downloaded from<br />
http://eurheartj.oxfordjournals.org/ by guest on June 7, 2013