Mutations in the mitochondrial thioredoxin reductase gene TXNRD2 ...
Mutations in the mitochondrial thioredoxin reductase gene TXNRD2 ...
Mutations in the mitochondrial thioredoxin reductase gene TXNRD2 ...
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
1126<br />
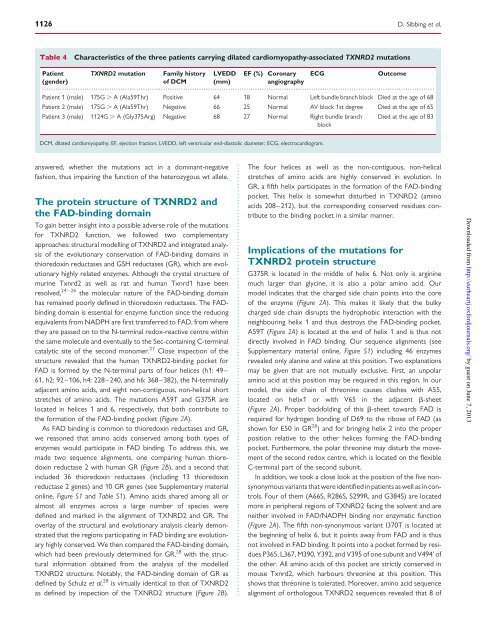
Table 4 Characteristics of <strong>the</strong> three patients carry<strong>in</strong>g dilated cardiomyopathy-associated <strong>TXNRD2</strong> mutations<br />
Patient <strong>TXNRD2</strong> mutation Family history LVEDD EF (%) Coronary ECG Outcome<br />
(gender)<br />
of DCM (mm)<br />
angiography<br />
...............................................................................................................................................................................<br />
Patient 1 (male) 175G . A (Ala59Thr) Positive 64 18 Normal Left bundle branch block Died at <strong>the</strong> age of 68<br />
Patient 2 (male) 175G . A (Ala59Thr) Negative 66 25 Normal AV block 1st degree Died at <strong>the</strong> age of 65<br />
Patient 3 (male) 1124G . A (Gly375Arg) Negative 68 27 Normal Right bundle branch<br />
block<br />
Died at <strong>the</strong> age of 83<br />
DCM, dilated cardiomyopathy; EF, ejection fraction; LVEDD, left ventricular end-diastolic diameter; ECG, electrocardiogram.<br />
answered, whe<strong>the</strong>r <strong>the</strong> mutations act <strong>in</strong> a dom<strong>in</strong>ant-negative<br />
fashion, thus impair<strong>in</strong>g <strong>the</strong> function of <strong>the</strong> heterozygous wt allele.<br />
The prote<strong>in</strong> structure of <strong>TXNRD2</strong> and<br />
<strong>the</strong> FAD-b<strong>in</strong>d<strong>in</strong>g doma<strong>in</strong><br />
To ga<strong>in</strong> better <strong>in</strong>sight <strong>in</strong>to a possible adverse role of <strong>the</strong> mutations<br />
for <strong>TXNRD2</strong> function, we followed two complementary<br />
approaches: structural modell<strong>in</strong>g of <strong>TXNRD2</strong> and <strong>in</strong>tegrated analysis<br />
of <strong>the</strong> evolutionary conservation of FAD-b<strong>in</strong>d<strong>in</strong>g doma<strong>in</strong>s <strong>in</strong><br />
thioredox<strong>in</strong> <strong>reductase</strong>s and GSH <strong>reductase</strong>s (GR), which are evolutionary<br />
highly related enzymes. Although <strong>the</strong> crystal structure of<br />
mur<strong>in</strong>e Txnrd2 as well as rat and human Txnrd1 have been<br />
resolved, 24 – 26 <strong>the</strong> molecular nature of <strong>the</strong> FAD-b<strong>in</strong>d<strong>in</strong>g doma<strong>in</strong><br />
has rema<strong>in</strong>ed poorly def<strong>in</strong>ed <strong>in</strong> thioredox<strong>in</strong> <strong>reductase</strong>s. The FADb<strong>in</strong>d<strong>in</strong>g<br />
doma<strong>in</strong> is essential for enzyme function s<strong>in</strong>ce <strong>the</strong> reduc<strong>in</strong>g<br />
equivalents from NADPH are first transferred to FAD, from where<br />
<strong>the</strong>y are passed on to <strong>the</strong> N-term<strong>in</strong>al redox-reactive centre with<strong>in</strong><br />
<strong>the</strong> same molecule and eventually to <strong>the</strong> Sec-conta<strong>in</strong><strong>in</strong>g C-term<strong>in</strong>al<br />
catalytic site of <strong>the</strong> second monomer. 27 Close <strong>in</strong>spection of <strong>the</strong><br />
structure revealed that <strong>the</strong> human <strong>TXNRD2</strong>-b<strong>in</strong>d<strong>in</strong>g pocket for<br />
FAD is formed by <strong>the</strong> N-term<strong>in</strong>al parts of four helices (h1: 49–<br />
61, h2: 92–106, h4: 228–240, and h6: 368–382), <strong>the</strong> N-term<strong>in</strong>ally<br />
adjacent am<strong>in</strong>o acids, and eight non-contiguous, non-helical short<br />
stretches of am<strong>in</strong>o acids. The mutations A59T and G375R are<br />
located <strong>in</strong> helices 1 and 6, respectively, that both contribute to<br />
<strong>the</strong> formation of <strong>the</strong> FAD-b<strong>in</strong>d<strong>in</strong>g pocket (Figure 2A).<br />
As FAD b<strong>in</strong>d<strong>in</strong>g is common to thioredox<strong>in</strong> <strong>reductase</strong>s and GR,<br />
we reasoned that am<strong>in</strong>o acids conserved among both types of<br />
enzymes would participate <strong>in</strong> FAD b<strong>in</strong>d<strong>in</strong>g. To address this, we<br />
made two sequence alignments, one compar<strong>in</strong>g human thioredox<strong>in</strong><br />
<strong>reductase</strong> 2 with human GR (Figure 2B), and a second that<br />
<strong>in</strong>cluded 36 thioredox<strong>in</strong> <strong>reductase</strong>s (<strong>in</strong>clud<strong>in</strong>g 13 thioredox<strong>in</strong><br />
<strong>reductase</strong> 2 <strong>gene</strong>s) and 10 GR <strong>gene</strong>s (see Supplementary material<br />
onl<strong>in</strong>e, Figure S1 and Table S1). Am<strong>in</strong>o acids shared among all or<br />
almost all enzymes across a large number of species were<br />
def<strong>in</strong>ed and marked <strong>in</strong> <strong>the</strong> alignment of <strong>TXNRD2</strong> and GR. The<br />
overlay of <strong>the</strong> structural and evolutionary analysis clearly demonstrated<br />
that <strong>the</strong> regions participat<strong>in</strong>g <strong>in</strong> FAD b<strong>in</strong>d<strong>in</strong>g are evolutionary<br />
highly conserved. We <strong>the</strong>n compared <strong>the</strong> FAD-b<strong>in</strong>d<strong>in</strong>g doma<strong>in</strong>,<br />
which had been previously determ<strong>in</strong>ed for GR, 28 with <strong>the</strong> structural<br />
<strong>in</strong>formation obta<strong>in</strong>ed from <strong>the</strong> analysis of <strong>the</strong> modelled<br />
<strong>TXNRD2</strong> structure. Notably, <strong>the</strong> FAD-b<strong>in</strong>d<strong>in</strong>g doma<strong>in</strong> of GR as<br />
def<strong>in</strong>ed by Schulz et al. 28 is virtually identical to that of <strong>TXNRD2</strong><br />
as def<strong>in</strong>ed by <strong>in</strong>spection of <strong>the</strong> <strong>TXNRD2</strong> structure (Figure 2B).<br />
The four helices as well as <strong>the</strong> non-contiguous, non-helical<br />
stretches of am<strong>in</strong>o acids are highly conserved <strong>in</strong> evolution. In<br />
GR, a fifth helix participates <strong>in</strong> <strong>the</strong> formation of <strong>the</strong> FAD-b<strong>in</strong>d<strong>in</strong>g<br />
pocket. This helix is somewhat disturbed <strong>in</strong> <strong>TXNRD2</strong> (am<strong>in</strong>o<br />
acids 208–212), but <strong>the</strong> correspond<strong>in</strong>g conserved residues contribute<br />
to <strong>the</strong> b<strong>in</strong>d<strong>in</strong>g pocket <strong>in</strong> a similar manner.<br />
Implications of <strong>the</strong> mutations for<br />
<strong>TXNRD2</strong> prote<strong>in</strong> structure<br />
D. Sibb<strong>in</strong>g et al.<br />
G375R is located <strong>in</strong> <strong>the</strong> middle of helix 6. Not only is arg<strong>in</strong><strong>in</strong>e<br />
much larger than glyc<strong>in</strong>e, it is also a polar am<strong>in</strong>o acid. Our<br />
model <strong>in</strong>dicates that <strong>the</strong> charged side cha<strong>in</strong> po<strong>in</strong>ts <strong>in</strong>to <strong>the</strong> core<br />
of <strong>the</strong> enzyme (Figure 2A). This makes it likely that <strong>the</strong> bulky<br />
charged side cha<strong>in</strong> disrupts <strong>the</strong> hydrophobic <strong>in</strong>teraction with <strong>the</strong><br />
neighbour<strong>in</strong>g helix 1 and thus destroys <strong>the</strong> FAD-b<strong>in</strong>d<strong>in</strong>g pocket.<br />
A59T (Figure 2A) is located at <strong>the</strong> end of helix 1 and is thus not<br />
directly <strong>in</strong>volved <strong>in</strong> FAD b<strong>in</strong>d<strong>in</strong>g. Our sequence alignments (see<br />
Supplementary material onl<strong>in</strong>e, Figure S1) <strong>in</strong>clud<strong>in</strong>g 46 enzymes<br />
revealed only alan<strong>in</strong>e and val<strong>in</strong>e at this position. Two explanations<br />
may be given that are not mutually exclusive. First, an unpolar<br />
am<strong>in</strong>o acid at this position may be required <strong>in</strong> this region. In our<br />
model, <strong>the</strong> side cha<strong>in</strong> of threon<strong>in</strong>e causes clashes with A55,<br />
located on helix1 or with V65 <strong>in</strong> <strong>the</strong> adjacent b-sheet<br />
(Figure 2A). Proper backfold<strong>in</strong>g of this b-sheet towards FAD is<br />
required for hydrogen bond<strong>in</strong>g of D69 to <strong>the</strong> ribose of FAD (as<br />
shown for E50 <strong>in</strong> GR 28 ) and for br<strong>in</strong>g<strong>in</strong>g helix 2 <strong>in</strong>to <strong>the</strong> proper<br />
position relative to <strong>the</strong> o<strong>the</strong>r helices form<strong>in</strong>g <strong>the</strong> FAD-b<strong>in</strong>d<strong>in</strong>g<br />
pocket. Fur<strong>the</strong>rmore, <strong>the</strong> polar threon<strong>in</strong>e may disturb <strong>the</strong> movement<br />
of <strong>the</strong> second redox centre, which is located on <strong>the</strong> flexible<br />
C-term<strong>in</strong>al part of <strong>the</strong> second subunit.<br />
In addition, we took a close look at <strong>the</strong> position of <strong>the</strong> five nonsynonymous<br />
variants that were identified <strong>in</strong> patients as well as <strong>in</strong> controls.<br />
Four of <strong>the</strong>m (A66S, R286S, S299R, and G384S) are located<br />
more <strong>in</strong> peripheral regions of <strong>TXNRD2</strong> fac<strong>in</strong>g <strong>the</strong> solvent and are<br />
nei<strong>the</strong>r <strong>in</strong>volved <strong>in</strong> FAD/NADPH b<strong>in</strong>d<strong>in</strong>g nor enzymatic function<br />
(Figure 2A). The fifth non-synonymous variant I370T is located at<br />
<strong>the</strong> beg<strong>in</strong>n<strong>in</strong>g of helix 6, but it po<strong>in</strong>ts away from FAD and is thus<br />
not <strong>in</strong>volved <strong>in</strong> FAD b<strong>in</strong>d<strong>in</strong>g. It po<strong>in</strong>ts <strong>in</strong>to a pocket formed by residues<br />
P365, L367, M390, Y392, and V395 of one subunit and V494’ of<br />
<strong>the</strong> o<strong>the</strong>r. All am<strong>in</strong>o acids of this pocket are strictly conserved <strong>in</strong><br />
mouse Txnrd2, which harbours threon<strong>in</strong>e at this position. This<br />
shows that threon<strong>in</strong>e is tolerated. Moreover, am<strong>in</strong>o acid sequence<br />
alignment of orthologous <strong>TXNRD2</strong> sequences revealed that 8 of<br />
Downloaded from<br />
http://eurheartj.oxfordjournals.org/ by guest on June 7, 2013