IMP Research Report 2002
IMP Research Report 2002
IMP Research Report 2002
You also want an ePaper? Increase the reach of your titles
YUMPU automatically turns print PDFs into web optimized ePapers that Google loves.
Figure 1: Eco1p - an acetyltransferase in cohesion. The image is an<br />
artist’s view of how our understanding of protein structure and<br />
function is emerging from a sequence alignment. It shows the joint<br />
multiple alignment of Eco1 sequences from different organisms and<br />
sequences of members of the GNAT superfamily of acetyltransferases.<br />
The alignment was obtained based on domain fragment<br />
searches, secondary structure predictions and physico-chemical<br />
similarity of amino acid types. The three-dimensional structure of<br />
GCN5 histone N-acetyltransferase from Tetrahymena thermophilum<br />
[Rojas et al. Nature 1999; 401:93-98] is shown above. The<br />
secondary structure elements of the most conserved motifs D, A,<br />
and B that were predicted in Eco1 are colored. Eco1 is a protein<br />
that has been previously implicated in the establishment of bridges<br />
between sister chromatids during DNA replication. The unexpected<br />
acetyltransferase activity of Eco1 is reported in the paper by D.<br />
Ivanov et al. (Curr Biol. <strong>2002</strong>, 12(4):323-328).<br />
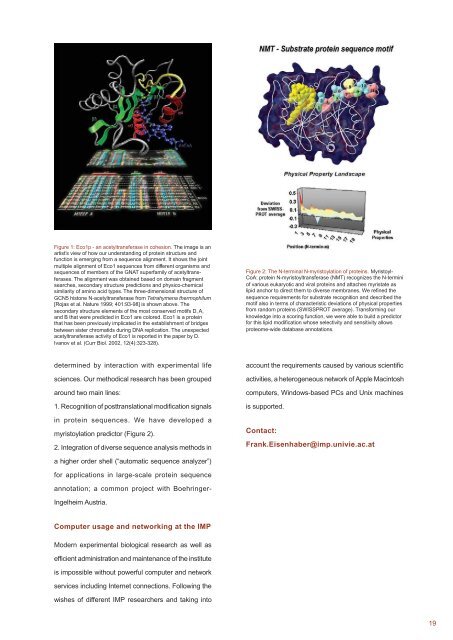
Figure 2: The N-terminal N-myristoylation of proteins. Myristoyl-<br />
CoA: protein N-myristoyltransferase (NMT) recognizes the N-termini<br />
of various eukaryotic and viral proteins and attaches myristate as<br />
lipid anchor to direct them to diverse membranes. We refined the<br />
sequence requirements for substrate recognition and described the<br />
motif also in terms of characteristic deviations of physical properties<br />
from random proteins (SWISSPROT average). Transforming our<br />
knowledge into a scoring function, we were able to build a predictor<br />
for this lipid modification whose selectivity and sensitivity allows<br />
proteome-wide database annotations.<br />
determined by interaction with experimental life<br />
sciences. Our methodical research has been grouped<br />
around two main lines:<br />
1. Recognition of posttranslational modification signals<br />
in protein sequences. We have developed a<br />
myristoylation predictor (Figure 2).<br />
2. Integration of diverse sequence analysis methods in<br />
account the requirements caused by various scientific<br />
activities, a heterogeneous network of Apple Macintosh<br />
computers, Windows-based PCs and Unix machines<br />
is supported.<br />
Contact:<br />
Frank.Eisenhaber@imp.univie.ac.at<br />
a higher order shell (“automatic sequence analyzer”)<br />
for applications in large-scale protein sequence<br />
annotation; a common project with Boehringer-<br />
Ingelheim Austria.<br />
Computer usage and networking at the <strong>IMP</strong><br />
Modern experimental biological research as well as<br />
efficient administration and maintenance of the institute<br />
is impossible without powerful computer and network<br />
services including Internet connections. Following the<br />
wishes of different <strong>IMP</strong> researchers and taking into<br />
19